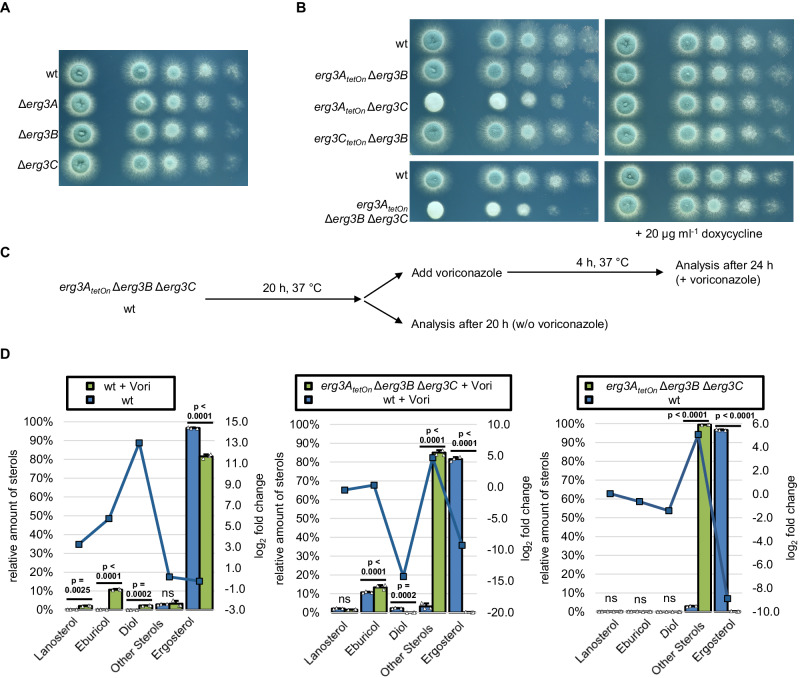
Fig. 5. Construction of a conditional sterol C5-desaturase mutant repression and the impact of sterol C5-desaturase depletion on the sterol pattern of azole-treated hyphae.
A, B In a series of tenfold dilutions derived from a starting suspension of 5 × 107 conidia ml−1 of wild type (wt) and the indicated sterol C5-desaturase mutant single, double, and triple mutants, aliquots of 3 µl were spotted onto AMM agar plates. AMM was supplemented with 20 μg ml−1 doxycycline when indicated. Agar plates were incubated at 37 °C, representative photos were taken after 28 h. C, D Conidia of wild type and the conditional sterol C5-desaturase mutant (erg3AtetOn Δerg3B Δerg3C) were inoculated in the Sabouraud medium. After 20 h incubation in a rotary shake at 37 °C, mycelium was either directly harvested or supplemented with 2 µg ml−1 voriconazole (+Vori), incubated in a rotary shake at 37 °C for another 4 h, and then harvested. For each condition, three biological replicates were cultured. D The sterol patterns of the harvested mycelia were analyzed by gas chromatography-mass spectrometry (GC-MS). The column graphs show the relative amounts (percentage of total sterol, left y-axis) of the indicated sterols for the indicated strains under the indicated conditions. The data points with the square symbols indicate the log2-fold change (right y-axis) in the amount of the respective sterol of the pairwise comparison of the conditions shown in the individual graphs. The log2-fold change data points were connected by lines for a better visual illustration of the changes in the profiles. Each column bar represents the mean of three replicates (data points) per condition, the error bars indicate standard deviations. Data were representative of three experiments conducted with two independent conditional ERG3 triple mutants under similar conditions. Statistical significance was set at p < 0.05, and calculated with a two-way ANOVA with Tukey’s multiple comparison test. p values are indicated in the graphs; ns not significant. Source data are provided as a Source Data file.