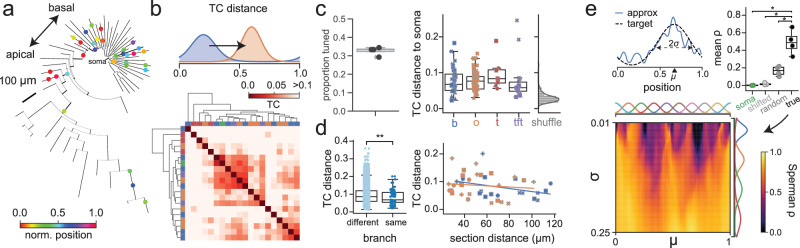
Fig. 5. Spatial tuning in non-place pyramidal cell dendrites.
a Example phylogram of reconstructed cell (AN061420-1), with recording locations colored by tuning curve centroid. b Example tuning curve distance (TCD, i.e. Wasserstein distance) matrix. Many segments exhibit high distance from soma. Top: Schematic of TCD calculation. TCD is normalized such that d = 1 corresponds to point masses on opposite sides of the belt. c Left: Proportion of tuned dendrites by cell, where tuning is defined as having a p-value below the 5th percentile of a random shuffle. Right: Cotuning of tuned dendrites with their respective soma. Black: N = 89 tuned dendrites from 4 cells; gray: TC distance 95th percentile from shuffle performed on each dendrite. Basal: p = 2 × 10−5, oblique: p = 8 × 10−9, trunk: p = 0.008, tuft: p = 0.02, Mann–Whitney U test. d Dendrites operate as compartments on the single-branch level. Left: Pairs of segments recorded on the same branch (n = 96 pairs) are significantly more co-tuned than pairs on different branches (n = 6568 pairs) (p = 0.0009, Mann-Whitney U test). e Arbitrary somatic tuning curves can be realized as sparse nonnegative combinations of tuned dendrites in non-place cells (n = 4 cells). Top left: Example tuning curve and approximation as weighted sum of dendrites. Top right: Mean realizability (Spearman’s ρ) of tuning curves in (bottom) using somatic tuning curve alone, somatic tuning curve with local shifts, and random tuning curves (true vs soma-only: p = 0.018, true vs soma/shifted: p = 0.007, true vs random: p = 0.006, paired t-test). Bottom: Realizability of theoretical Gaussian tuning curves parameterized by location and width as nonnegative least-squares fit from tuned dendrites. Unless otherwise specified, all box-and-whisker plots show Q1, Q2, Q3, and range excluding outliers. All statistical tests performed in a two-sided manner where relevant unless otherwise specified.