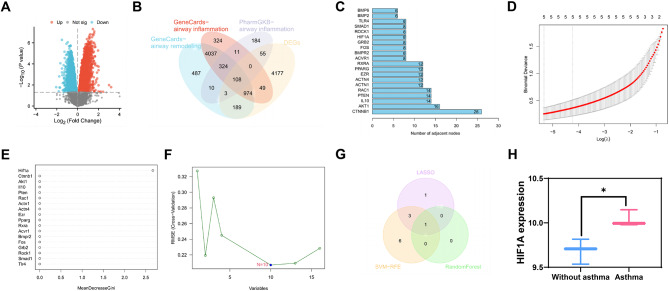
Fig. 1.
A machine learning algorithm for screening mRNA related to airway remodeling and inflammation. Note (A) Volcano plot of differentially expressed mRNA between Asthma group and Without asthma group in high-throughput sequencing data of mouse lung tissues; (B) Venn diagram showing the intersection of significantly differentially expressed mRNA in Asthma mouse lung tissues in sequencing data, mRNA related to airway remodeling and inflammation in the Genecards database, and mRNA related to airway inflammation in the PharmGKB database; (C) Statistics of the number of adjacent nodes of core genes in the gene interaction network graph, where the x-axis represents the number of adjacent nodes and the y-axis represents gene names; (D) LASSO regression coefficient selection plot; (E) Random forest algorithm result plot; (F) SVM-RFE analysis result plot; (G) Venn diagram showing the intersection of Asthma-related mRNA selected by three machine learning algorithms: LASSO regression, random forest algorithm, and SVM-RFE; (H) Expression of HIF-1α in the sequencing data, displaying the results as the logarithmic values of gene expression (with 3 cases without asthma and 3 cases with asthma). In the volcano plot, blue dots represent significantly downregulated mRNA in the Asthma group, red dots represent significantly upregulated mRNA in the Asthma group, and gray dots represent mRNA with no significant difference. Note * indicates P < 0.05 compared to the Without asthma group