FIGURE 1.
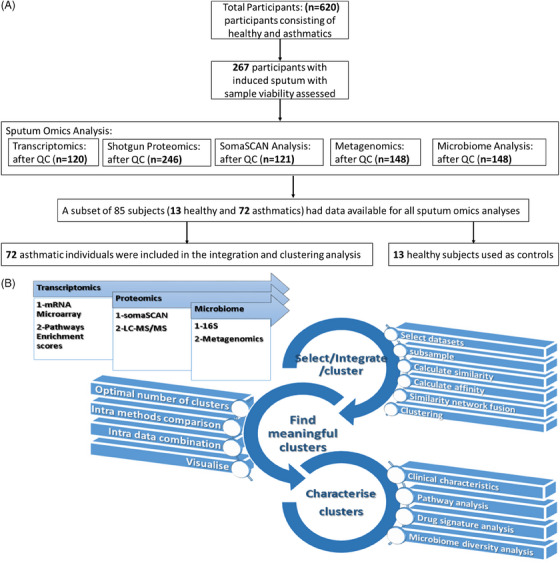
Machine learning workflow. (A) CONSORT flow chart reporting the patients and controls we studied. (B) Three types of data were the starting point of the workflow, that is, transcriptomics, proteomics and microbiome date. Each of these three data types included 2 data matrices either derived by incorporating available knowledge or from various bioanalytical platforms, for example, proteomics data from LC‐MS/MS and somaSCAN, microbiome from 16S and Metagenomics platforms. The workflow consists of three main multi‐faceted compartments: (1) select/integrate/cluster, (2) find meaningful clusters, (3) characterise clusters. The first compartment is about running a data integration and clustering algorithm on different combination of data and generating clusters. The second compartment includes multiple steps to calculate, compare and visualise various groupings generated by the first compartment. The goal of this step is to aid decision making about the optimal number of clusters. Finally, the third compartment characterises the only clustering result that is deemed to be most suitable and stable through the second compartment.
