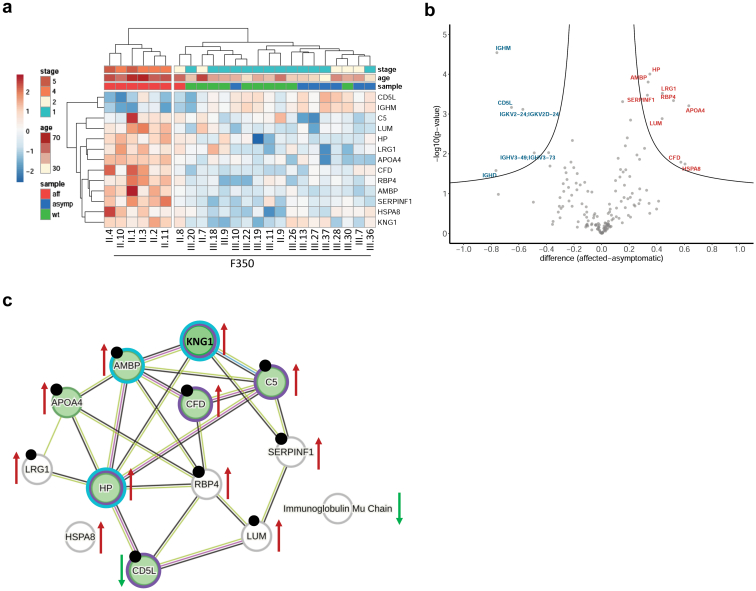
Figure 8.
Glycoproteomic analysis of plasma. (a) Unsupervised hierarchical clustering of 12 glycoproteins identified by mass spectrometry and analysis of variance with significantly different abundance between samples of genetically affected individuals with CKD (aff), genetically affected individuals with normal kidney function CKD (asymp) and genetically unaffected family members (wt) from F350. Values are shown as Z-scores, and together with CKD stage and age of participants, are presented as the color codes, which scale is shown in the corresponding lookup tables. (b) Volcano plot demonstrating differences in plasma proteins abundancies identified by mass spectrometry between genetically affected individuals with CKD (aff) and genetically affected individuals with normal kidney function CKD (asymp). The X-axis shows the difference as a log2 value of a ratio of Z-scores of individual proteins; the Y-axis shows the statistical significance of the difference. (c) Physical interactions of proteins with significantly different abundance between genetically affected individuals with CKD and genetically affected individuals with normal kidney function CKD visualized by STRING database. Color circles depict proteins associated with immune response (Violet - DISGENET database), proteins associated with kidney disease, kidney insufficiency or kidney injury (Turquoise - DISGENET database), proteins associated with hemostasis (Red - STRING database), proteins associated with complement and coagulation cascade (Green – STRING database), proteins which were not associated with any of the listed category in STRING or DISGENET database (Gray) and proteins associated with either stage of CKD or progression of CKD or ESKD from the literature (Black dots). Arrows indicate an increase or decrease in corresponding protein abundance in genetically affected individuals with CKD. CKD, chronic kidney disease.