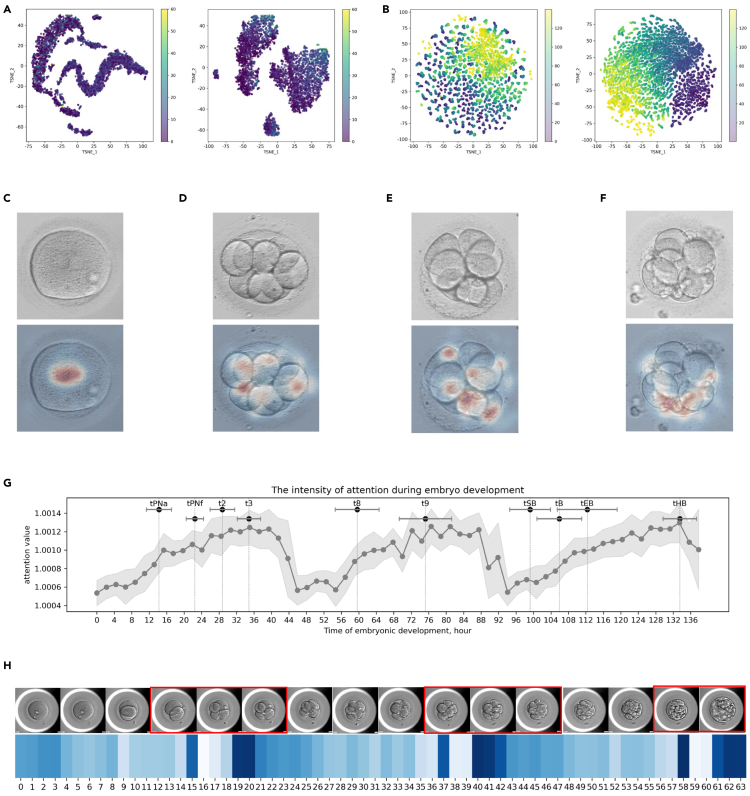
Figure 4.
Visualization of evidence for embryo morphology and ploidy assessment
(A and B) Visualization of embryo representations learned by VTCLR via t-SNE. Representations were extracted from (A) embryo images and (B) frames of time-lapse videos in the pre-training dataset. Each point is colored according to its corresponding label of (A) embryo fragmentation (%) and (B) hours post insemination (HPI) on time-lapse video (hours).
(C–F) Visualization of evidence for embryo morphological assessment using the integrated gradients method. The original image (upper) and an attention map generated from our model are presented. Upper panels: the original embryo images. Lower panels: explanation method-generated saliency heatmaps. (C) Normal pronuclear type of day 1 (good); (D) blastomere cell number of day-3 embryo (normal); (E) blastomere symmetry of day-3 embryo (good); (F) fragmentation rate of day-3 embryo (normal).
(G) Average intensity of attention generated by AI during embryo development. The intensity values of attention were extracted from the pre-trained temporal encoder with all frames averaged over cases in the internal validation set. Each bar on the top represents the mean and standard deviation values of HPI of a defined time-lapse morphokinetic parameter.
(H) Case study of the temporal attention for embryo ploidy at specific frames during embryo development. Blue-colored maps indicate the attention generated by the temporal encoder of the given time-lapse video. The peaks in these maps corresponded to the temporal location of morphokinetic characteristics, which are highlighted in red boxes.