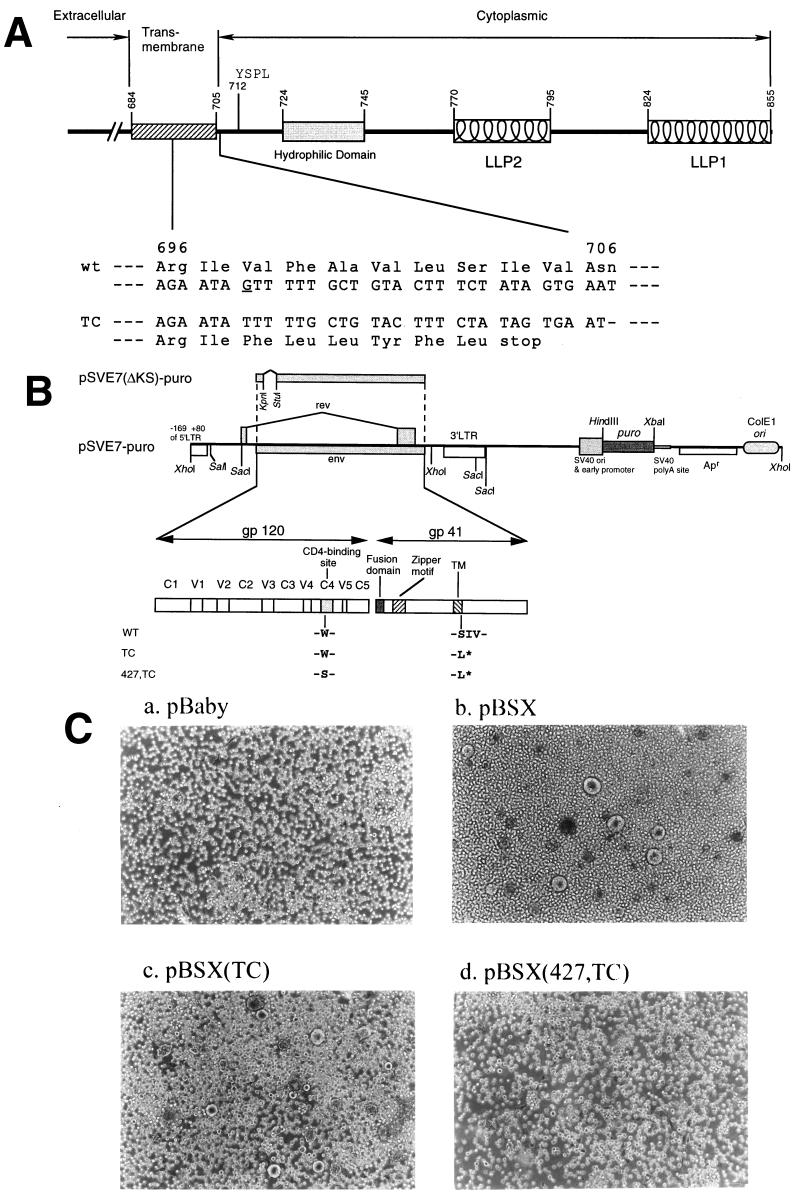
FIG. 1.
Construction and characterization of Env mutants. (A) A schematic representation of the cytoplasmic domain truncation Env mutant. Structural motifs in gp41 cytoplasmic domains are shown, such as internalization signal YSPL and the tyrosine-based basolateral-targeting signal located at residue 712, a highly hydrophilic region, and two positively charged amphipathic α-helices marked LLP-1 and LLP-2. The amino acid residues are numbered according to their positions in the Env of the HXB2 strain. The region encompassing residues from 696 to 706 and its corresponding DNA sequence are indicated. The deletion of the G base as underlined located at nucleotide 8315 of the HXB2 sequence results in a frame shift of the open reading frame and a premature termination of translation after leucine at position 703 in the TM domain. (B) Construction of wt and mutant pSVE7-puro plasmids. A puromycin resistance expression cassette was inserted into the wt pSVE7. The KpnI-BamHI fragments isolated from various mutant pSVE7 plasmids were substituted for the corresponding sequence in wt pSVE7-puro to generate various mutant pSVE7-puro plasmids. The asterisk represents the stop codon in the TM domain as described in panel A. Single-letter amino acid codes are used. (C) Analysis of the syncytium-forming ability of Env mutants. COS-1 cells were transfected with 2 μg each of pBaby, wt, or mutant pBSX as indicated. Two days after transfection, 106 SupT1 cells were added into each transfected culture, and photographs were taken 18 h after coculture under a light microscope. Magnification, ×100.