FIGURE 1.
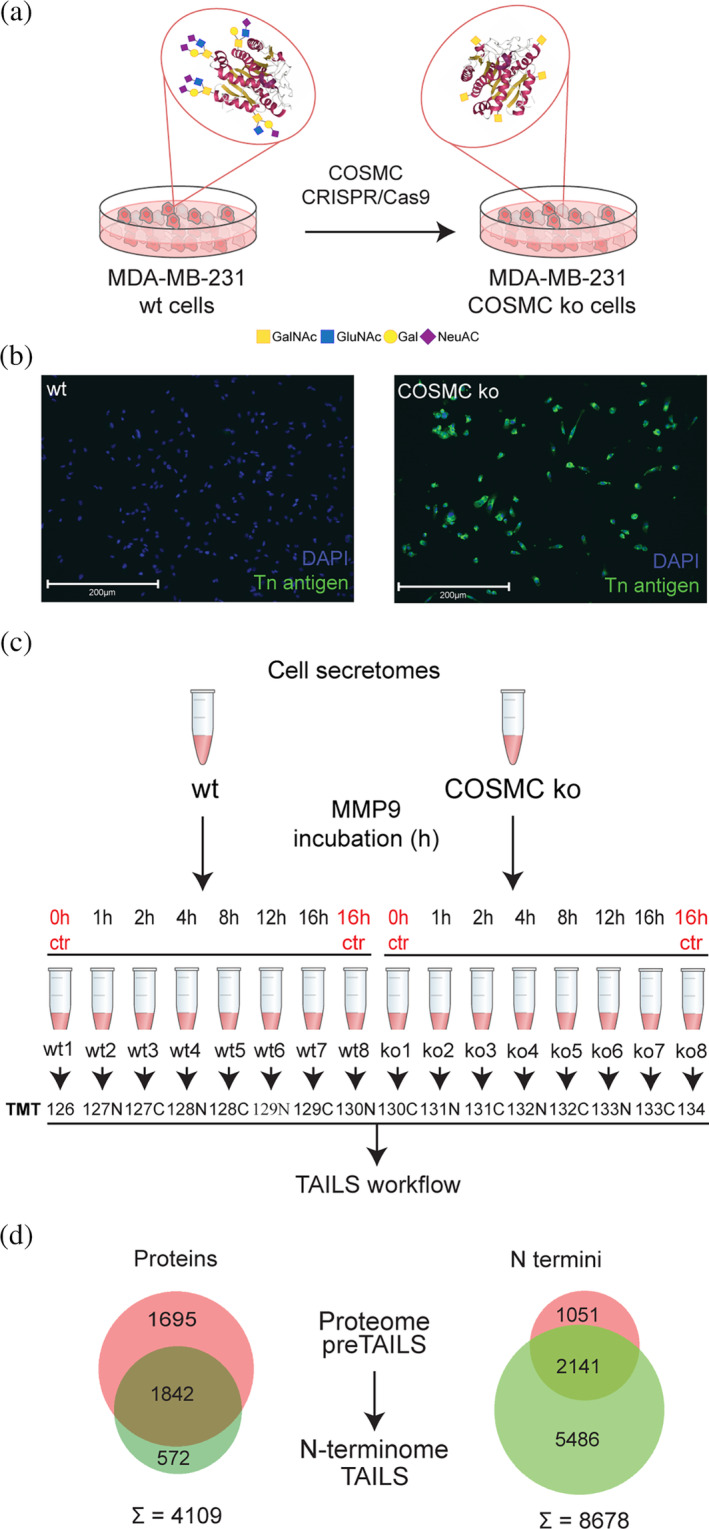
Proteome and N‐terminome analysis of wt and COSMC ko MDA‐MB‐231 cells. (a) COSMC was genetically inactivated by CRISPR/Cas9 to generate MDA‐MB‐231 COSMC ko with truncated O‐glycan trees. GalNAc, N‐acetyl‐d‐galactosamine; GlcNAc, N‐acetyl‐d‐glucosamine; Gal, d‐galactose; NeuAc, N‐acetylneuraminic acid. (b) Immunofluorescence analysis of Tn antigen confirmed functional inactivation in COSMC KO cells. DAPI was used to visualize cell nuclei. Scale bar: 200 μm. N = 2. (c) Cell secretomes were collected from wt and COSMC ko MDA‐MB‐231 cells, incubated with active recombinant human MMP9 for increasing time points, and analyzed by 16plex‐TMT‐TAILS. Individual samples from the time‐series experiments were treated as replicates in quantitative comparisons between genotypes. (d) Venn diagram illustrating the distribution and overlap of the identified 4109 proteins and 8678 N termini between the preTAILS and TAILS samples.
