Figure 2.
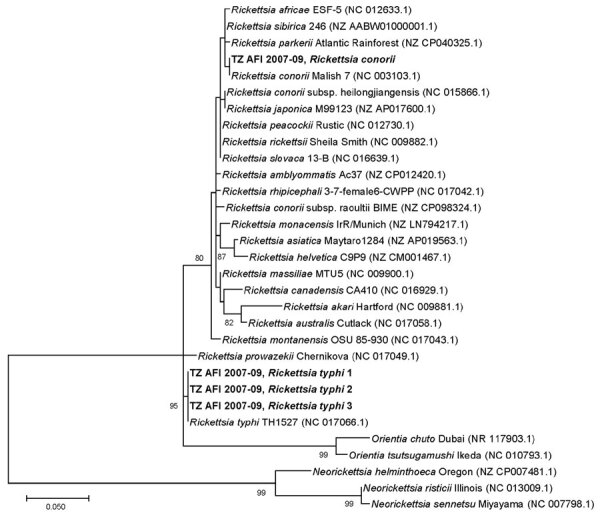
Phylogenetic tree of Rickettsia spp. sequences detected in metagenomic analysis of bacterial zoonotic pathogens among febrile patients, Tanzania, 2007–2009. The tree compares sequences from the 16S variable regions 1 and 2 (V1–V2) of the Rickettsia cohort from this study (bold text) to sequences from closely related Rickettsia species. Numbers in parentheses indicate GenBank accession numbers. The sequence from the study sample with R. conorii aligned 100% R. conorii strain Malish (accession no. NC003103.1) and was distinct from R. africae (accession no. NC012633.1). All 3 R. typhi strains from this study aligned 100% with R. typhi reference strain (accession no. NC017066.1) and were distinct from R. prowazekii (accession no. NC017049.1). The V1–V2 16S target is not sufficient to differentiate between R. conorii subsp. heilogjiangensis and R. japonica, or between R. rickettsia, R. peacockii, R. philipii, and R. slovaca. However, the V1–V2 16S target is sufficient to differentiate between R. conorii conorii and R. africae because 2 single-neucleotide differences would be expected between R. conorii conorii and R. africae and a TTT insertion in R. africae. Scale bar indicates nucleotide subsitutions per site.
