Abstract
Leptospirosis is a common but underdiagnosed zoonosis. We conducted a 1-year prospective study in La Guaira State, Venezuela, analyzing 71 hospitalized patients who had possible leptospirosis and sampling local rodents and dairy cows. Leptospira rrs gene PCR test results were positive in blood or urine samples from 37/71 patients. Leptospira spp. were isolated from cultured blood or urine samples of 36/71 patients; 29 had L. interrogans, 3 L. noguchii, and 4 L. venezuelensis. Conjunctival suffusion was the most distinguishing clinical sign, many patients had liver involvement, and 8/30 patients with L. interrogans infections died. The Leptospira spp. found in humans were also isolated from local rodents; L. interrogans and L. venezuelensis were isolated from cows on a nearby, rodent-infested farm. Phylogenetic clustering of L. venezuelensis isolates suggested a recently expanded outbreak strain spread by rodents. Increased awareness of leptospirosis prevalence and rapid diagnostic tests are needed to improve patient outcomes.
Keywords: leptospirosis, Leptospira, Leptospira venezuelensis, Leptospira interrogans, liver disease, kidney disease, Weil’s disease, intermediate clade, intermediate species, cows, humans, bacteria, zoonoses, Venezuela
Leptospirosis, one of the most common zoonoses worldwide, (1,2) is caused by Leptospira spp. In humans, its most severe, multiorgan, potentially fatal form is known as Weil’s disease (3). Leptospira can also infect animals, such as cattle, sheep, cats, and dogs. Rodents are the reservoir for most Leptospira spp.; rodent kidneys can become colonized with Leptospira and chronically shed the bacteria in urine. Except for occupational or recreational exposure, leptospirosis generally occurs in residents of marginal, rodent-infested areas, often in coastal regions of tropical countries (3).
According to their ability to cause human disease, Leptospira bacteria were originally divided into fully pathogenic (P1), intermediate pathogenic (P2), and saprophytic or nonpathogenic (S1 and S2) subclades; this phylogenetic separation is confirmed by genome sequencing (4,5). The pathogenic species, most commonly L. interrogans, can cause leptospirosis and Weil’s disease, but the role of intermediate species in human illness is unclear (5). Intermediate Leptospira spp. have been discovered by environmental sampling of soil and water (5), but they have also been found in animals and humans, where they are thought to cause only mild, self-limited illness without liver, kidney, or pulmonary involvement (5).
Leptospira infections are classically diagnosed by using the microscopic agglutination test (MAT) to detect Leptospira-specific antibodies, but diagnosis often requires comparing titers of acute and convalescent serum samples. Culturing Leptospira for a definitive bacteriologic diagnosis is difficult and takes weeks to months. Therefore, Leptospira bacteria are usually detected by PCR of blood or urine samples and identified by sequencing the amplified genes and comparing those sequences to known Leptospira spp. (6).
Venezuela is considered a moderate-incidence country for leptospirosis (7), but the true incidence is unknown because of a lack of clinical recognition of the disease and difficulties in laboratory diagnosis. To determine the presence of Leptospira spp., identify local strains, and evaluate leptospirosis incidence in Venezuela, we performed a prospective study in La Guaira, a small state on Venezuela’s Caribbean coast. Although the study was conducted in 2010–2011 and reporting delayed because of Venezuela’s economic situation, we believe the clinical leptospirosis data and epigenomic study of an intermediate Leptospira sp. outbreak remain relevant.
Methods
Ethics Approval
The Bioethics Commission of the Instituto Venezolano de Investigaciones Científicas, Caracas, Venezuela, approved the human study. The National Office of Biologic Diversity within the Venezuela Ministry for the Environment (Document 0264) and the Instituto Venezolano de Investigaciones Científicas Commission on Animal Bioethics approved the capture of rodents.
Study Area
We included patients with possible leptospirosis in La Guaira State, located on the northern Caribbean coast of Venezuela. La Guaira contains a shipping port and the nation’s principal airport and has a population of ≈353,000. It is a beach resort for residents of Caracas but also contains low socioeconomic urban and rural areas. In the 1999 Vargas tragedy, mudslides destroyed much of the infrastructure of La Guaira (formerly Vargas State), causing thousands of fatalities.
Patient Selection
We visited Dr. José María Vargas Hospital during March 2010–March 2011 and Dr. Rafael Medina Jiménez Hospital during March–July 2010 and February–March 2011; visits were >2 times per week each. We reviewed diagnoses of new patients at admission and questioned hospital staff about new patients who had clinical symptoms suggestive of leptospirosis. Inclusion criteria were residence or place of work in La Guaira and an initial evaluation that included >1 sign or symptom of leptospirosis as described by the World Health Organization (8): fever >38°C with unknown etiology for <21 days, fever with renal failure (anuria, oligouria, or elevated creatinine), abdominal or muscle pain, icterus, conjunctival suffusion, hypokalemia or hyponatremia, hemoptysis or pulmonary hemorrhage, or an initial diagnosis of hepatitis or dengue. After patients voluntarily signed an informed consent form, we interviewed those patients and collected their clinical histories and places of residence. We also consulted the physician’s notes. We excluded patients who were unable to complete the interview or provide adequate data. We enrolled a total of 71 patients from whom blood and urine specimens were obtained. Of those 71 patients, 38 had serologic tests for dengue and 39 for hepatitis A or B. Frozen serum samples from some patients were subsequently tested for hepatitis viruses A and B by PCR (Appendix Table 1).
Leptospira Cultures
Leptospira were cultured at 28–30°C in liquid or semisolid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium with 10% supplement and 50–100 μg/mL of 5-fluorouracil for initial cultures (Appendix). All solutions and media were prepared according to the World Health Organization technical manual (8).
Rodent Capture
We set up Sherman aluminum traps in urban areas close to the residences of patients who were PCR positive for Leptospira (Appendix). The species of captured rodents were determined by amplifying and sequencing a subunit of the cytochrome c oxidase gene (9).
Cow Samples
We collected blood with and without EDTA anticoagulant from the caudal vein of 16 crossbred Bos taurus × Bos indicus (predominantly Bos taurus) dairy cows. Cows were 3–10 years of age and located on a farm in Miranda State, Venezuela, ≈30 km from La Guaira State (Appendix). We collected urine samples from the same cows after intramuscular injection of the diuretic furosemide (1 mg/kg). We cultured blood and urine samples and performed PCR for the Leptospira genes rrs (16S rDNA) and lipL32.
Passaging of Isolates in Hamsters
We intraperitoneally injected Leptospira isolates from second to fourth passages of liquid culture into 4-week-old male Syrian golden hamsters (Mesocricetus auratus). Sixteen days after injection, we euthanized the hamsters and removed and macerated the kidneys. We placed the kidney tissue into EMJH medium to sediment and then inoculated culture medium with the supernatant.
Molecular Detection of Leptospira
We amplified lipL32 (10) and regions V3–V6 of the rrs gene from isolated DNA by using PCR (11) (Appendix Table 1). We purified the rrs gene amplification products (QIAGEN, https://www.qiagen.com), which were then sequenced by Macrogen (https://www.macrogen.com). We also sequenced the lig gene from a few specimens (12). We used the L. interrogans genes pntA, sucA, pfkB, tpiA, mreA, glmU, and caiB (13,14) for multilocus sequence typing (MLST). We performed variable-number tandem-repeat (VNTR) analysis of L. interrogans isolates as previously described (15).
MAT of Bovine Serum Samples
MATs were performed in the bacteriology laboratory of the Instituto Nacional de Investigaciones Agricolas according to 2003 Pan American Health Organization standards (https://www.paho.org/es/documentos/leptospirosis-humana-guia-para-diagnostico-vigilancia-control). MATs were considered positive when >50% of Leptospira bacteria were agglutinated.
Phylogenetic Reconstruction of L. venezuelensis Isolates
We used Velvet (16) for de novo assembly of genome contigs from sequencing reads of L. venezuelensis isolates. We used cow isolate 201502610 (GenBank Biosample accession no. SAMEA5168082) as a reference to map reads from the other L. venezuelensis isolates (Appendix).
Statistics
We performed statistical analyses of patient signs and symptoms and clinical test values by using Stata 13 (StataCorp LLC, https://www.stata.com). We did not adjust p values for multiple statistical testing.
Results
PCR and Cultures of Patient Specimens
Through twice-weekly visits to the 2 hospitals in La Guaira state over a 1-year period, we identified 71 patients who met the inclusion criteria (Appendix Tables 2, 3). We PCR amplified the Leptospira rrs gene from blood samples of 17, urine samples of 22, and both blood and urine samples of 2 patients. We also cultured Leptospira bacteria from blood samples from 13, urine samples from 20, and both blood and urine samples from 3 patients (Appendix Table 4). Using PCR amplification of Leptospira rrs in either blood or urine samples as confirmation of leptospirosis, the sensitivity of the rrs gene for diagnosing leptospirosis was 46% for blood and 59% for urine specimens; both sample types had 100% specificity. For lipL32 PCR amplification, sensitivity was 41% for blood, 35% for urine, and 70% when both blood and urine samples were tested; all samples had 100% specificity. For blood cultures, sensitivity was 43%, and specificity was 100%; for urine cultures, sensitivity was 59%, and specificity was 97%; for either positive blood or urine cultures, sensitivity was 95%, and specificity was 97% (Table 1; Appendix Tables 4–6). The rrs gene was amplified from 2 patients who had negative Leptospira cultures: from a blood specimen of a patient with jaundice and from the urine of a patient who died of severe pulmonary disease.
Table 1. Tests used for leptospirosis diagnoses in study of outbreak of intermediate species Leptospira venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela*.
| Diagnostic test† | rrs PCR+, n = 37‡ | rrs PCR–, n = 34‡ | % Sensitivity (95% CI) | % Specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | % Accuracy (95% CI) |
|---|---|---|---|---|---|---|---|
| rrs PCR | |||||||
| Blood, + | 17 | 0 | 46 (30–63) | 100 (90–100) | 100 (80–100) | 63 (56–70) | 72 (60–82) |
| Blood, – | 20 | 34 | NA | NA | NA | NA | NA |
| Urine, + | 22 | 0 | 59 (42–75) | 100 (90–100) | 100 (85–100) | 69 (61–77) | 79 (68–88) |
| Urine, – | 15 | 34 | NA | NA | NA | NA | NA |
| Both, + |
2 |
0 |
NA |
NA |
NA |
NA |
NA |
| Culture | |||||||
| Blood, + | 16 | 0 | 43 (27–61) | 100 (90–100) | 100 (80–100) | 62 (55–68) | 70 (58–81) |
| Urine, + | 22 | 1 | 59 (42–75) | 97 (85–100) | 96 (76–99) | 69 (60–77) | 77 (66–87) |
| Either, + |
35 |
1 |
95 (82–99) |
97 (85–100) |
97 (84–100) |
94 (81–98) |
96 (88–99) |
| lipL32 PCR | |||||||
| Blood, + | 15 | 0 | 41 (25–58) | 100 (90–100) | 100 (78–100) | 61 (54–67) | 69 (57–79) |
| Urine, + | 13 | 0 | 35 (20–53) | 100 (90–100) | 100 (75–100) | 59 (53–64) | 66 (54–77) |
| Either, + | 26 | 0 | 70 (53–84) | 100 (90–100) | 100 (87–100) | 76 (65–84) | 85 (74–92) |
*Diagnostic values were obtained for Leptospira cultures and PCR of Leptospira lipL32 and Leptospira rrs (16S rDNA) genes of blood and urine specimens from hospitalized patients. NA, not applicable; NPV, negative predictive value; PPV, positive predictive value; –, negative; +, positive. †Test results are shown for patients who had Leptospira detected in blood, urine, or either blood or urine. ‡Total numbers of study patients who had the Leptospira rrs gene detected by PCR of either blood or urine samples.
Initial diagnoses were similar for patients in this study who had positive or negative Leptospira PCR and were most commonly dengue, hepatitis, icteric hemorrhagic syndrome, febrile syndrome, or unknown. Leptospirosis was listed as an initial diagnosis for 6 patients from whom Leptospira spp. were isolated and for 1 patient who had negative Leptospira cultures. Dengue was diagnosed in 3 patients and hepatitis in 4 patients who had positive Leptospira cultures; dengue was diagnosed in 5 patients and hepatitis in 4 patients who had negative cultures. Leptospira isolation was not correlated with seasonal variation in precipitation.
We compared PCR sequences of rrs with GenBank sequences by using BLAST (17). We identified 29 sequences as L. interrogans, 3 as L. noguchii, and 4 were 99% identical to the intermediate species L. licerasiae and L. wolffi (Appendix Tables 7, 8); genome sequencing showed those 4 isolates belonged to a novel intermediate species that we then named L. venezuelensis (18). The lig gene (12) was amplified by PCR from the urine of the culture-negative patient who died of pulmonary disease and was identified as belonging to L. interrogans by using BLAST. Patients who had positive tests for hepatitis or dengue and positive Leptospira blood or urine cultures all grew L. interrogans and were assumed to be co-infected. Serum samples from L. venezuelensis–positive patients were negative for hepatitis viruses A and B (Appendix Table 9).
Clinical Characteristics
Patients who had PCR-amplified rrs were more likely to have conjunctival suffusion, dyspnea, cough, hemoptysis, and myalgias (Figure 1; Appendix Tables 10, 11). Eight (27%) of the 30 patients who had L. interrogans infections died of their illness, whereas no deaths were recorded among the 34 patients who had no evidence of leptospirosis. Leptospirosis patients who died had more severe infections, with pulmonary and renal involvement, than did those who survived (Figure 2; Appendix Tables 12, 13). Patients who died of leptospirosis had more hemoptysis but less abdominal pain and myalgias and also had higher mean urea and creatinine levels, higher leukocyte counts, higher percentages of neutrophils, and lower percentages of lymphocytes than those who survived.
Figure 1.
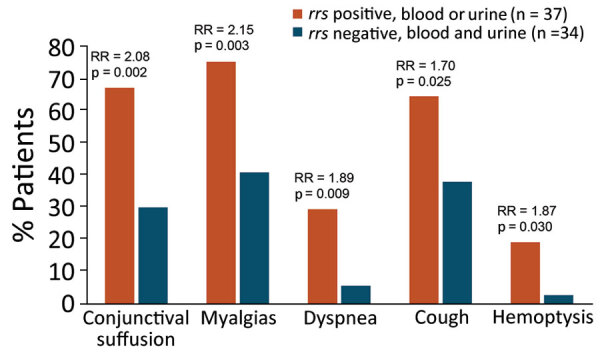
Distinguishing clinical features of hospitalized patients in study of outbreak of intermediate species Leptospira venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela. The most statistically different clinical symptoms are shown for hospitalized patients considered to have leptospirosis according to positive PCR for the Leptospria rrs gene in either blood or urine specimens compared with those without leptospirosis according to negative rrs PCR in both blood and urine samples. PCR primers for rrs amplify a region of the gene encoding 16S rRNA that is highly conserved in Leptospira (Appendix Table 1). One patient whose urine culture grew L. venezuelensis was rrs PCR negative, and leptospirosis was not diagnosed (Appendix Table 9). Comparisons of all clinical features with 95% CIs were also determined (Appendix Tables 10, 11). RRs and Pearson χ2 test p values were calculated by using Stata 13 (StataCorp LLC, https://www.stata.com). RR, risk ratio.
Figure 2.
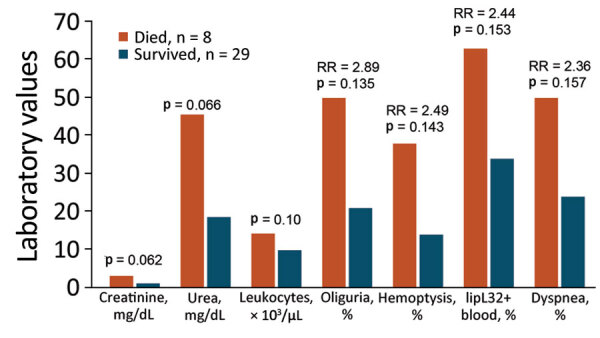
Clinical features most strongly associated with fatal outcomes in study of outbreak of intermediate species Leptospira venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela. Clinical features are shown for hospitalized patients who had positive PCR tests for the Leptospira rrs (16S rDNA) gene in blood or urine and either survived or succumbed to their illness. Laboratory units of measure are indicated on the x axis for each bar. Comparisons of all clinical features with 95% CIs were also determined (Appendix Tables 12, 13). p values comparing creatinine, urea, and number of lymphocytes were obtained from Pearson χ2 tests. p values comparing percentages of patients with oliguria, hemoptysis, lipL32, and dyspnea were obtained from 2-tailed t-tests. All statistical calculations were performed by using Stata 13 (StataCorp LLC, https://www.stata.com). RR, risk ratio.
Cultures from Captured Rodents
To delineate reservoir hosts for Leptospira, we captured 45 rodents from 27 communities where patients who had positive cultures resided. We captured 30 Mus musculus mice and 11 Rattus rattus and 4 R. norvegicus rats. We amplified the rrs gene by PCR and cultured Leptospira from kidney tissue samples from all 45 rodents; 36 (80%) isolates were L. interrogans, 4 (9%) were L. noguchii, 3 (7%) were the intermediate species L. fainei, and 2 (4%) were L. venezuelensis. L. interrogans was isolated from all 3 rodent species, L. noguchii was isolated only from mice, and the intermediate species L. fainei and L. venezuelensis were only isolated from R. rattus rats.
Leptospira in Cows on Nearby Farm
Leptospira spp. are known to infect cattle. In a preliminary study, we performed MATs on serum samples from 48 cows on 8 small farms in adjacent Miranda State. We found Leptospira-specific antibodies against >1 Leptospira serovars in 2 animals from a single farm located ≈30 km from where the leptospirosis patients in this study resided. We then obtained blood and urine specimens from 16 cows randomly selected from that single farm and performed MATs against live antigens of 23 Leptospira reference strains; 9 samples agglutinated >1 serovar (Table 2; Appendix Table 14). Of those 9 cows, 8 had urine positive for Leptospira rrs by PCR; 2 urine samples had positive cultures of L. interrogans, and 7 had positive cultures of L. venezuelensis. The cows that had L. venezuelensis–positive urine had MAT titers of 1:400 to 1:800 against reference strain L. interrogans serovar Wolffi, serogroup Sejroe. The 2 L. interrogans–positive cows had high MAT titers for other serovars: 1:400 for L. hebdomadis (cow 5) and 1:400 for L. mini (cow 9). Cows 1 and 8 were negative according to MATs and rrs PCR of their urine, but their blood samples were rrs PCR–positive. Urine of cow 8 grew L. interrogans, whereas urine of cow 1 grew L. venezuelensis (Table 2; Appendix Table 14). Cow 11 had a MAT titer of 1:1,600 for L. interrogans serovar Bataviae and rrs-positive urine, but no Leptospira spp. were isolated from either the blood or urine. L. venezuelensis isolates did not agglutinate with antiserum to common L. interrogans serovars, although antiserum to serovar Wolffi was not included.
Table 2. Analysis of blood and urine specimens from cows in study of outbreak of intermediate species Leptospira venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela* .
| Cow no. |
rrs PCR |
lipL32 PCR |
Serology |
Cultures |
Sequenced species† | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood | Urine | Blood | Urine | MAT titer | Serovar | Blood | Urine | |||||
| 1 | + | – | – | – | Negative | NA | + | – | L. venezuelensis | |||
| 2 | – | – | – | – | Negative | NA | – | – | NA | |||
| 3 | – | + | – | – | 1:800 | L. wolffii | – | + | L. venezuelensis | |||
| 4 | – | – | – | – | Negative | NA | – | – | NA | |||
| 5 | – | + | – | + | 1:400 | L. hebdomadis | – | + | L. interrogans | |||
| 6 | – | – | – | – | Negative | NA | – | – | NA | |||
| 7 | – | + | – | – | 1:800 | L. wolffii | – | + | L. venezuelensis | |||
| 8 | + | – | + | – | Negative | NA | + | – | L. interrogans | |||
| 9 | – | + | – | + | 1:400 | L. mini | – | + | L. interrogans | |||
| 10 | – | – | – | – | Negative | NA | – | – | NA | |||
| 11 | – | + | – | – | 1:1,600 | L. bataviae | – | – | NA | |||
| 12 | – | + | – | – | 1:800 | L. wolffii | – | + | L. venezuelensis | |||
| 13 | – | + | – | – | 1:400 | L. wolffii | – | + | L. venezuelensis | |||
| 14 | – | + | – | – | 1:800 | L. wolffii | – | + | L. venezuelensis | |||
| 15 | – | – | – | – | Negative | NA | – | – | NA | |||
| 16 | – | + | – | – | 1:400 | L. wolffii | – | + | L. venezuelensis | |||
*Results for MAT serology, cultures, and PCR of the Leptospira lipL32 and rrs (16S rDNA) genes from cultured isolates. MAT, microscopic agglutination test; NA, not applicable; –, negative; +, positive. †Leptospira spp. were determined by sequencing the PCR-amplified rrs gene.
Growth in Hamsters
We purified all Leptospira isolates by injecting early passage cultures into the peritoneal cavities of Syrian golden hamsters and performing necropsies 16 days after inoculation; 3 hamsters infected with L. interrogans and 1 infected with L. noguchii died before 16 days. We cultured aliquots of macerated kidney extracts from all inoculated hamsters and performed PCR to detect Leptospira rrs. In each case, sequences of rrs from the hamster kidney extracts were identical to the sequences from the corresponding original specimens and also the Leptospira cultured from those hamster kidney extracts.
Molecular Epidemiology of L. interrogans
Among the L. interrogans strains isolated from humans or rodents, 3 clusters had 7/7 identical MLST alleles; in each cluster, 2 patients resided in the same residential zone (Table 3). Of the 27 different MLST profiles, only 4 were present in the Leptospira PubMLST database (https://pubmlst.org/organisms/leptospira-spp), 2 of which (sequence types 27 and 50) were in clusters that had 7/7 identical alleles. Sequence types 20 and 37 were clustered with strains that had 6/7 identical alleles. VNTR clustering was not concordant with MLST clustering (Appendix Table 15). Two of the 3 L. interrogans strains isolated from cows had identical alleles in 4 VNTR loci (Appendix Table 16) but were not analyzed by using MLST.
Table 3. MLST of Leptospira interrogans isolates in study of outbreak of intermediate species L. venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela*.
| Isolates† | MLST allele nos. |
ST‡ | ||||||
|---|---|---|---|---|---|---|---|---|
| glmU | pntA | sucA | tpiA | pfkB | mreA | caiB | ||
| Human | ||||||||
| CAB-H41 | 1 | 1 | 2 | 1 | 7 | 7 | 8 | NP* |
| CAY-U48 | 1 | 1 | 2 | 1 | 7 | 4 | 3 | 20 |
| CAB-U03 | 1 | 1 | 2 | 2 | 7 | 4 | 3 | NP |
| MAC-H04 | 1 | 1 | 2 | 2 | 7 | 4 | 5 | NP |
| URI-U06 | 1 | 1 | 3 | 2 | 7 | 4 | 3 | NP |
| URI-H01 | 1 | 1 | 3 | 2 | 7 | 4 | 3 | NP |
| CLM-H09 | 1 | 1 | 3 | 2 | 4 | 7 | 5 | NP |
| CLM-U30 | 1 | 3 | 2 | 2 | 4 | 4 | 19 | NP |
| MAC-H63 | 1 | 3 | 2 | 2 | 7 | 7 | 19 | NP |
| CAY-H65 | 1 | 3 | 3 | 1 | 4 | 5 | 5 | NP |
| SOB-U13 | 1 | 12 | 3 | 3 | 10 | 4 | 5 | NP |
| MAQ-U18 | 1 | 12 | 3 | 3 | 10 | 5 | 19 | NP |
| CLM-U22 | 1 | 12 | 2 | 3 | 10 | 6 | 19 | NP |
| CLM-U28 | 1 | 12 | 3 | 3 | 10 | 6 | 19 | 27 |
| CLM-H08 | 1 | 12 | 3 | 3 | 10 | 6 | 19 | 27 |
| GUA-H40 | 1 | 12 | 3 | 3 | 10 | 6 | 19 | 27 |
| CLM-U45 | 3 | 3 | 3 | 2 | 4 | 5 | 5 | NP |
| CLM-U47 | 3 | 3 | 3 | 3 | 4 | 5 | 5 | 37 |
| NAG-U02 | 6 | 1 | 3 | 2 | 4 | 7 | 3 | NP |
| CAY-U49 | 6 | 1 | 3 | 3 | 76 | 7 | 3 | NP |
| CLM-U46 | 6 | 2 | 3 | 3 | 7 | 7 | 19 | NP |
| CLM-U24 | 6 | 1 | 3 | 12 | 4 | 5 | 5 | NP |
| GUA-H52 | 6 | 3 | 2 | 2 | 4 | 4 | 3 | NP |
| GUA-H64 | 6 | 3 | 2 | 3 | 4 | 7 | 5 | NP |
| CAB-U11 | 6 | 3 | 3 | 2 | 4 | 5 | 5 | NP |
| GUA-H21 | 6 | 3 | 3 | 3 | 1 | 7 | 5 | NP |
| CAO-U23 | 6 | 3 | 3 | 3 | 4 | 5 | 19 | NP |
| MAQ-H53 | 6 | 8 | 2 | 2 | 9 | 7 | 5 | 50 |
|
MAQ-H60
|
6
|
8
|
2
|
2
|
9
|
7
|
5
|
50 |
| Rat | ||||||||
| CLM-R09-A | 1 | 1 | 2 | 2 | 7 | 4 | 8 | NP |
| CLM-R11-A | 1 | 1 | 3 | 3 | 4 | 6 | 19 | NP |
| SOB-R13-B§ | 1 | 12 | 3 | 3 | 10 | 6 | 19 | 27 |
*Bold font indicates isolates that had identical profiles. MLST, multilocus sequence typing; NP, not present in database; ST, sequence type. †The first 3 letters for each isolate indicate the area of the patient’s residence or where the rodent was captured in the state of La Guaira, Venezuela: CAB, Caraballeda; CAO, Caruao; CAY, Carayaca; CLM, Catia La Mar; GUA, La Guaira; MAC, Macuto; MAQ, Maiquetia; NAG, Niguata; SOB, Soublette; or URI, Urimare. The fourth letter is H (isolated from human blood), U (isolated from human urine), or R (isolated from rat tissue). ‡STs found in the Leptospira PubMLST database (https://pubmlst.org/organisms/leptospira-spp) (14). §Rat sequence shared an MLST profile for some alleles with human isolates CLM-U28, CLM-H08, and GUA-H40.
New Intermediate Species of Leptospira
L. venezuelensis, isolated from 4 patients (Appendix Table 9), 2 rodents, and 7 cows, is located on the phylogenetic tree within the Leptospira intermediate pathogen or P2 subclade (5). Three of the 4 patients infected with L. venezuelensis resided in the same municipality; the fourth patient resided in an adjacent district. This municipality was the most frequent residence of leptospirosis patients, home to 10 of 32 patients with other Leptospira infections. Of the 2 rats infected with L. venezuelensis, 1 was trapped in the same municipality and the other in a nearby district.
Phylogenetic reconstruction of genomes from the 6 sequenced L. venezuelensis isolates uncovered limited genetic diversity (Figure 3). The isolates from human, rodent, and bovine hosts all differed by <12 single-nucleotide polymorphisms (SNPs), suggesting a recent outbreak of a L. venezuelensis strain that was spread, presumably by rodents, to different host populations. The L. venezuelensis genome sequence data have been deposited in GenBank (Biosample accession nos. SAMEA5168082, SAMEA5168083, SAMEA5168130, SAMEA5168133, SAMEA5168318, SAMN06855518, SAMN39993761, SAMN39993762, and SAMN39993763).
Figure 3.
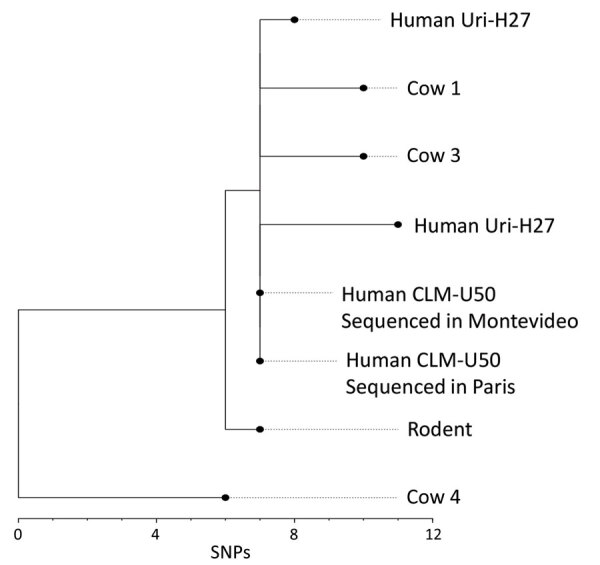
Phylogenetic analysis of Leptospira venezuelensis isolates in study of outbreak of intermediate species L. venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela. Branch length indicates the number of SNPs separating L. venezuelensis strains. Phylogenetic tree was reconstructed according to comparisons of whole-genome sequences from 6 L. venezuelensis strains isolated from hospitalized leptospirosis patients in La Guaira State on the Caribbean coast of Venezuela, from rodents captured near the residences of hospitalized leptospirosis patients, and from dairy cows on a farm 30 km away from La Guaira State. Human isolate CLM-50 was sequenced at both the Institute Pasteur in Paris, France, and the Institute Pasteur in Montevideo, Uruguay. Human isolate Uri-H27 was sequenced twice at the Institute Pasteur in Paris; the genome of the isolate after many passages in culture contained 3 SNPs that were not present in the same isolate from an earlier passage. Scale bar indicates number of SNPs per site. SNP, single-nucleotide polymorphism.
Discussion
The true incidence of leptospirosis in La Guaira state has been unknown, likely because it has been difficult or impossible to diagnose and has not been considered by clinicians, even in patients with characteristic signs and symptoms. Our prospective search for leptospirosis cases in La Guaira’s 2 hospitals during a 1-year period found rrs PCR evidence of Leptospira spp. in blood or urine specimens from 37 hospitalized patients, including 8 patients who died. We also cultured Leptospira from 36 patient samples. Two patients with positive rrs PCR had negative cultures, but 1 of those patients had an L. interrogans lig gene fragment amplified from their urine. The population of La Guaira is ≈353,000, corresponding to a borderline high incidence of 10 leptospirosis cases/100,000 population. However, this figure is almost certainly an underestimate because the study only included patients ill enough to require hospitalization and did not capture patients with less severe illness, who represent up to 90% of leptospirosis cases (2).
Leptospira spp. were isolated from the kidneys of all 45 rodents captured in the region. Leptospira species distributions were similar in rodents and humans; most isolates were L. interrogans, which is globally the species most associated with severe human illness. L. venezuelensis was also isolated from 7 cows on a nearby farm, whereas L. interrogans was isolated from only 3 cows on the same farm (Table 2; Appendix Table 14).
Leptospirosis is difficult to diagnose in a clinically useful time frame, but rrs PCR of both blood and urine samples detected 37 cases. The most discriminative clinical finding in patients was conjunctival suffusion (19), but Leptospira-positive patients also had more myalgias, dyspnea, cough, and hemoptysis than did hospitalized Leptospira-negative patients. L. interrogans was recovered from patients with the most severe cases, and 27% (8/30) of L. interrogans–infected patients died. However, for patients with mild to moderate disease, the infecting species could not be distinguished by patient signs, symptoms, or laboratory values (Appendix Table 8).
The intermediate species L. fainei was isolated from the kidneys of 3/45 captured rodents. L. fainei has been reported to cause disease in humans (20) but was not isolated from any human patient or bovid in this study. L. venezuelensis is phylogenetically closer to other intermediate species reported to cause human illness, such as L. liceraciae (21) and L. wolffi (22) and is phylogenetically close to Leptospira spp. isolated from environmental samples in Malaysia, Mayotte, and New Caledonia (5).
Few studies have been conducted to determine the phylogenetic relatedness of different strains of Leptospira spp. isolated from a particular geographic region. L. interrogans isolates from this study had many MLST profiles, including clusters of profiles found in the Leptospira PubMLST database (Table 3). MATs showed that serum samples from 3 cows each reacted to a different L. interrogans serovar, including 2 whose isolates had the same VNTR pattern (Appendix Tables 14, 16). The heterogeneity of L. interrogans strains suggests a long-term endemic presence in the local rodent population. In contrast, the genomes of 6 L. venezuelensis isolates differed by a maximum of 11 SNPs (Figure 3), suggesting an outbreak strain. Although only 6 of the 13 L. venezuelensis isolates were sequenced, they were obtained from a diverse sampling of hospitalized humans, rats captured in La Guaira, and cows on a farm 30 km away from patient residences. Unless L. venezuelensis has a mutation rate even slower than slow-mutating Mycobacterium tuberculosis (23), the low genetic diversity reflects a recently expanded bacteria population. Greater genomic heterogeneity would be expected if L. venezuelensis evolved from a local environmental Leptospira sp. Instead, the close genomic similarity between isolates suggests a recent introduction of L. venezuelensis into the region, perhaps arriving with rats on a ship that docked in the port of La Guaira and then spread within the local rodent population.
Infections with intermediate clade Leptospira spp. have only rarely been associated with icteric human illness (6), but 3 of 4 patients from whom L. venezuelensis was isolated were icteric, had liver aminotransferase values >250 (Appendix Table 9) and negative test results for hepatitis viruses A and B. Only 1 patient with L. venezuelensis infection was tested for dengue, but all 4 had platelet levels within reference ranges, which is uncharacteristic for acute dengue.
Although intermediate Leptospira spp. are thought to be incapable of surviving in an animal model, infection of rats has been reported for the intermediate species L. licerasiae (24). We recovered all 13 L. venezuelensis isolates from hamster kidneys 16 days after intraperitoneal inoculation of low passage isolates, although later passages of the same isolates could not be recovered from hamsters after high-dose intraperitoneal infections (data not shown). The acquisition of SNPs and loss of virulence during in vitro passages of Leptospira isolates has been previously described (25,26).
Leptospira intermediate species are often isolated from environmental samples (5), but it seems unlikely that L. venezuelensis was an environmental or laboratory contaminant. The rrs PCR of the original human, bovine, and rodent specimens; the isolate cultures; and hamster infection studies were all performed separately before sequencing results were available, and the samples containing L. venezuelensis were not temporally linked. The MAT titers of serum samples from L. venezuelensis–positive bovids all showed the same presumed cross-reaction with L. interrogans serovar Wolffi, consistent with the genomic evidence of an outbreak strain. Human disease causality could be confirmed by high or rising MAT titers in patient serum samples, but acute serum samples from 2 L. venezuelensis and 4 L. interrogans patients did not have titers >1:50, and convalescent patient blood samples were not collected.
In Argentina (27), L. wolffii was isolated from a patient who died of a severe respiratory syndrome, but PCR results suggested an L. interrogans co-infection. Similarly, 2 of the 4 L. venezuelensis–positive patients in this study had positive lipL32 PCR results (Appendix Table 9). The lipL32 primers were designed to amplify lipL32 from L. interrogans or other pathogenic Leptospira spp. but not from intermediate species, such as L. venezuelensis. Although the amplified lipL32 fragments were not sequenced, the 2 lipL3-positive patients could have been co-infected with L. venezuelensis and L. interrogans. Another patient from whom L. venezuelensis was cultured had negative rrs PCR results in both blood and urine. The pathogenicity of this intermediate species could not be confidently evaluated from the 4 L. venezuelensis–positive patients in this study.
In conclusion, an L. venezuelensis outbreak circulating in rodents appears to have spread to cows in the region and also infected humans, in whom it might have caused febrile illness with hepatic involvement. Our findings indicate the need for increased awareness of leptospirosis prevalence and characteristics in Venezuela and other tropical, rodent infested coastal regions and also indicates an urgent need for rapid point-of-care tests to diagnose leptospirosis and improve patient treatment and outcomes.
Additional information for outbreak of intermediate species Leptospira venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela.
Acknowledgments
We thank Albert Ko, Paula Ristow, and the staff of the Centro de Referencia Internacional de Leptospirosis de la Fundación Oswaldo Cruz, Salvador de Bahía, Brazil, for training L.C. and generously providing reference strains; Raphael Puche and the Unidad de Estudios Genéticos y Forenses (Instituto Venezolano de Investigaciones Científica) for genomic sequencing and analysis; Benjamin Valencia for help in verifying patient data; Mathieu Picardeau and his laboratory for genomic sequencing, hamster infections, serovar tests, help, and advice; Hans van der Linden and Harry Bannister for help interpreting the MAT results; and Yadira Castillo, Yurimia Oropeza, Rumania Miranda, Alexandra González, Mariela Gómez, Carmen Bogado, and Miguel Damargo for help identifying patients, collecting biological samples, and capturing rodents. Without their kind and generous assistance, this study would not have been possible.
This work was supported by the regular budget allotment of the Instituto Venezolano de Investigaciones Científicas to the Laboratorio de Genética Molecular and Institut Pasteur (project no. PTR 30-17).
Biography
Ms. Caraballo received her undergraduate degree in biology from the University of Zulia, Venezuela, and is pursuing a PhD in the Laboratorio de Genética Molecular, Centro de Microbiología y Biología Celular, Instituto Venezolano de Investigaciones Científica, Caracas, Venezuela. Her principal research interest is the molecular epidemiology of human and animal leptospirosis.
Footnotes
Suggested citation for this article: Caraballo L, Rangel Y, Reyna-Bello A, Muñoz M, Figueroa-Espinosa R, Sanz-Rodriguez CE, et al. Outbreak of intermediate species Leptospira venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela. Emerg Infect Dis. 2024 Aug [date cited]. https://doi.org/10.3201/eid3008.231562
Current affiliation: Universidad de las Fuerzas Armadas ESPE, Santo Domingo, Ecuador.
Current affiliation: Universidad de Buenos Aires, Buenos Aires, Argentina.
Current affiliation: Institut Pasteur de Montevideo, Montevideo, Uruguay.
Current affiliation: The University of North Carolina, Chapel Hill, North Carolina, USA.
References
- 1.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis. 2015;9:e0003898. 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Stobart Gallagher MA, Dunn N. Leptospirosis. In: StatPearls. Treasure Island (FL): StatPearls Publishing; 2024. [PubMed] [Google Scholar]
- 3.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. ; Peru-United States Leptospirosis Consortium. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–71. 10.1016/S1473-3099(03)00830-2 [DOI] [PubMed] [Google Scholar]
- 4.Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, Berg DE, et al. What makes a bacterial species pathogenic?: comparative genomic analysis of the genus Leptospira. PLoS Negl Trop Dis. 2016;10:e0004403. 10.1371/journal.pntd.0004403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl Trop Dis. 2019;13:e0007270. 10.1371/journal.pntd.0007270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balamurugan V, Gangadhar NL, Mohandoss N, Thirumalesh SRA, Dhar M, Shome R, et al. Characterization of leptospira isolates from animals and humans: phylogenetic analysis identifies the prevalence of intermediate species in India. Springerplus. 2013;2:362. 10.1186/2193-1801-2-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12:351–7. 10.1016/j.ijid.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. Human leptospirosis: guidance for diagnosis, surveillance and control. 2003. [cited 2023 Oct 15]. https://www.who.int/publications/i/item/human-leptospirosis-guidance-for-diagnosis-surveillance-and-control
- 9.Robins JH, McLenachan PA, Phillips MJ, Craig L, Ross HA, Matisoo-Smith E. Dating of divergences within the Rattus genus phylogeny using whole mitochondrial genomes. Mol Phylogenet Evol. 2008;49:460–6. 10.1016/j.ympev.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 10.Bomfim MRQ, Koury MC. Evaluation of LSSP-PCR for identification of Leptospira spp. in urine samples of cattle with clinical suspicion of leptospirosis. Vet Microbiol. 2006;118:278–88. 10.1016/j.vetmic.2006.07.020 [DOI] [PubMed] [Google Scholar]
- 11.Ahmed A, Anthony RM, Hartskeerl RA. A simple and rapid molecular method for Leptospira species identification. Infect Genet Evol. 2010;10:955–62. 10.1016/j.meegid.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 12.Palaniappan RUM, Chang YF, Chang CF, Pan MJ, Yang CW, Harpending P, et al. Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol Cell Probes. 2005;19:111–7. 10.1016/j.mcp.2004.10.002 [DOI] [PubMed] [Google Scholar]
- 13.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. 10.1371/journal.pntd.0000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Bailey MS, Holden MTG, et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl Trop Dis. 2013;7:e1954. 10.1371/journal.pntd.0001954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salaün L, Mérien F, Gurianova S, Baranton G, Picardeau M. Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J Clin Microbiol. 2006;44:3954–62. 10.1128/JCM.00336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010;11:11.5. [DOI] [PMC free article] [PubMed]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 18.Puche R, Ferrés I, Caraballo L, Rangel Y, Picardeau M, Takiff H, et al. Leptospira venezuelensis sp. nov., a new member of the intermediate group isolated from rodents, cattle and humans. Int J Syst Evol Microbiol. 2018;68:513–7. 10.1099/ijsem.0.002528 [DOI] [PubMed] [Google Scholar]
- 19.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arzouni JP, Parola P, La Scola B, Postic D, Brouqui P, Raoult D. Human infection caused by Leptospira fainei. Emerg Infect Dis. 2002;8:865–8. 10.3201/eid0808.010445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthias MA, Ricaldi JN, Cespedes M, Diaz MM, Galloway RL, Saito M, et al. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis. 2008;2:e213. 10.1371/journal.pntd.0000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman S, Paul SK, Aung MS, Ahmed S, Haque N, Raisul MNI, et al. Predominance of Leptospira wolffii in north-central Bangladesh, 2019. New Microbes New Infect. 2020;38:100765. 10.1016/j.nmni.2020.100765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet. 2012;13:601–12. 10.1038/nrg3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez C, Lubar AA, Vinetz JM, Matthias MA. Experimental infection of Rattus norvegicus by the group II intermediate pathogen, Leptospira licerasiae. Am J Trop Med Hyg. 2018;99:275–80. 10.4269/ajtmh.17-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picardeau M. Toolbox of molecular techniques for studying Leptospira spp. Curr Top Microbiol Immunol. 2018;415:141–62. 10.1007/82_2017_45 [DOI] [PubMed] [Google Scholar]
- 26.Lehmann JS, Corey VC, Ricaldi JN, Vinetz JM, Winzeler EA, Matthias MA. Whole genome shotgun sequencing shows selection on Leptospira regulatory proteins during in vitro culture attenuation. Am J Trop Med Hyg. 2016;94:302–13. 10.4269/ajtmh.15-0401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiani Y, Jacob P, Varni V, Landolt N, Schmeling MF, Pujato N, et al. Isolation and clinical sample typing of human leptospirosis cases in Argentina. Infect Genet Evol. 2016;37:245–51. 10.1016/j.meegid.2015.11.033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information for outbreak of intermediate species Leptospira venezuelensis spread by rodents to cows and humans in L. interrogans–endemic region, Venezuela.


