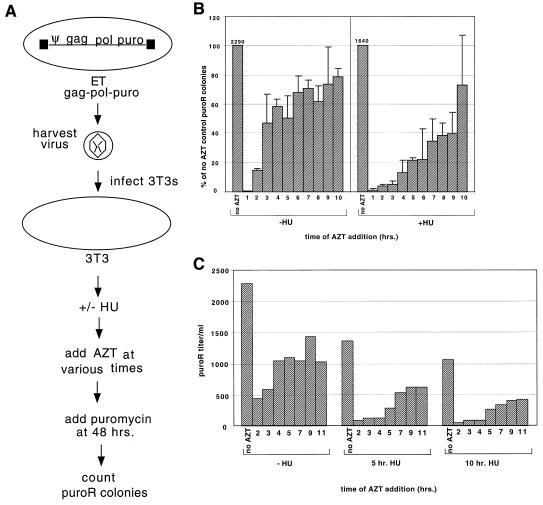
FIG. 1.
Assay of timing of reverse transcription. (A) Schematic overview of timing of reverse transcription assays. ET cells stably expressing gag-pol-puro were used as a source of virus for the timing assays. (ET cells are 293T cells that stably express ecotropic Env.) 3T3 cells were infected for 10 min with ET gag-pol-puro virus, and the cells were washed to remove most unattached virions. After being washed, cells were treated with 60 μM HU for 5 h or remained untreated. At various times postinfection, 200 μM AZT was added to individual plates. Puromycin was added to the plates 48 h postinfection. Colonies were counted after 2 weeks of selection. (B) Averaged results of two independent experiments measuring timing of reverse transcription. The left half of the graph shows results of experiments carried out in the absence of HU (−HU). The positive control received no AZT, and the negative control (data not shown) had no virus or AZT and no colonies were present. Numbers 1–10 denote the times of AZT addition (hours postinfection). The right half of the graph shows results of experiments done in the presence of 60 μM HU (+HU). The positive control received HU but no AZT. For these experiments, the media on all plates were changed at 5 h postinfection. The average puromycin-resistant (puroR) colony titers for positive controls in experiments conducted in the absence and presence of HU were 2,290 and 1,640 colonies/ml, respectively, as indicated above the bars. (C) Timing of reverse transcription with varying times of HU exposure. Cells were infected as described for panel B except that they were treated with 60 μM HU for 5 or 10 h or remained untreated (−HU). At various times postinfection (2, 3, 4, 5, 7, 9, or 11 h) 200 μM AZT was added to individual plates. AZT was removed and puromycin was added after 48 h, and colonies were counted 2 weeks later.