FIGURE 3.
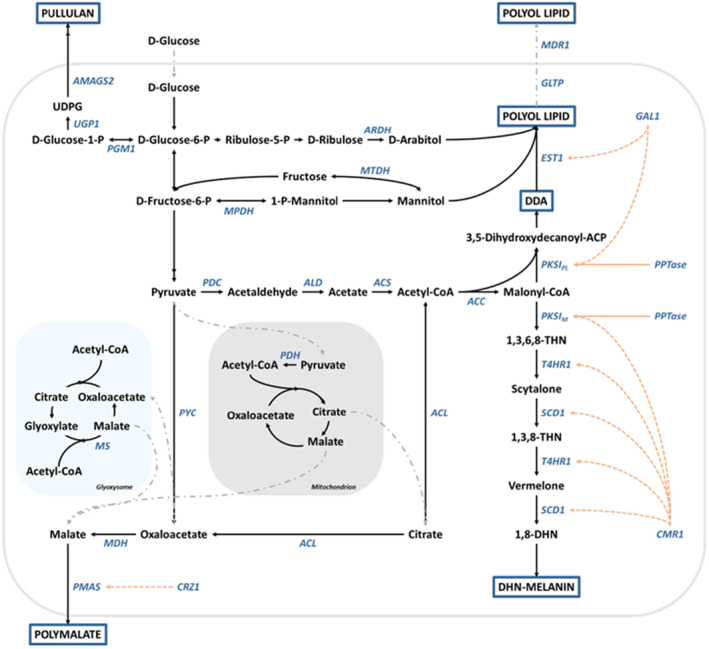
The biosynthetic pathways for pullulan, polyol lipids, polymalate, and DHN‐melanin in the genus Aureobasidium. Genes coding for the respective enzymes are shown in blue. 1,3,6,8‐THN, 1,3,6,8‐tetrahydroxynaphtalene; 1,3,8‐THN, 1,3,8‐trihydroxynaphtalene; 1,8‐DHN, 1,8‐dihydroxynaphtalene; ACC, acetyl‐CoA carboxylase; ACL, ATP‐citrate lyase; ACP, acyl carrier protein; ACS, acetyl‐CoA synthetase; ALD, acetaldehyde dehydrogenase; AMAGS2, a multidomain α‐glucan synthetase; ARDH, arabitol dehydrogenase; ATP, adenosine triphosphate; CMR1, transcription factor; CoA, coenzyme A; CRZ1, transcription factor; DDA, oligo‐dihydroxydecanoic acid; EST1, esterase 1; GAL1, transcription factor; GLTP, glycolipid transfer protein; MDH, malate dehydrogenase; MDR1, ATP‐binding cassette transporter; MPDH, mannitol‐1‐phosphate dehydrogenase; MS, malate synthase; MTDH, mannitol dehydrogenase; P, phosphate; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase; PGM1, phosphoglucomutase 1; PKSIM, polyketide synthase type I involved in melanin biosynthesis; PKSIPL, polyketide synthase type I involved in polyol lipid biosynthesis; PMAS, polymalate synthetase; PPTase, phosphopantetheinyl transferase; PYC, pyruvate carboxylase; SCD1, scytalone dehydratase 1; T4HR1, 1,3,6,8‐THN/1,3,8‐THN reductase; UDPG, uridine diphosphate glucose; UGP1, UDP‐glucose pyrophospholase.
