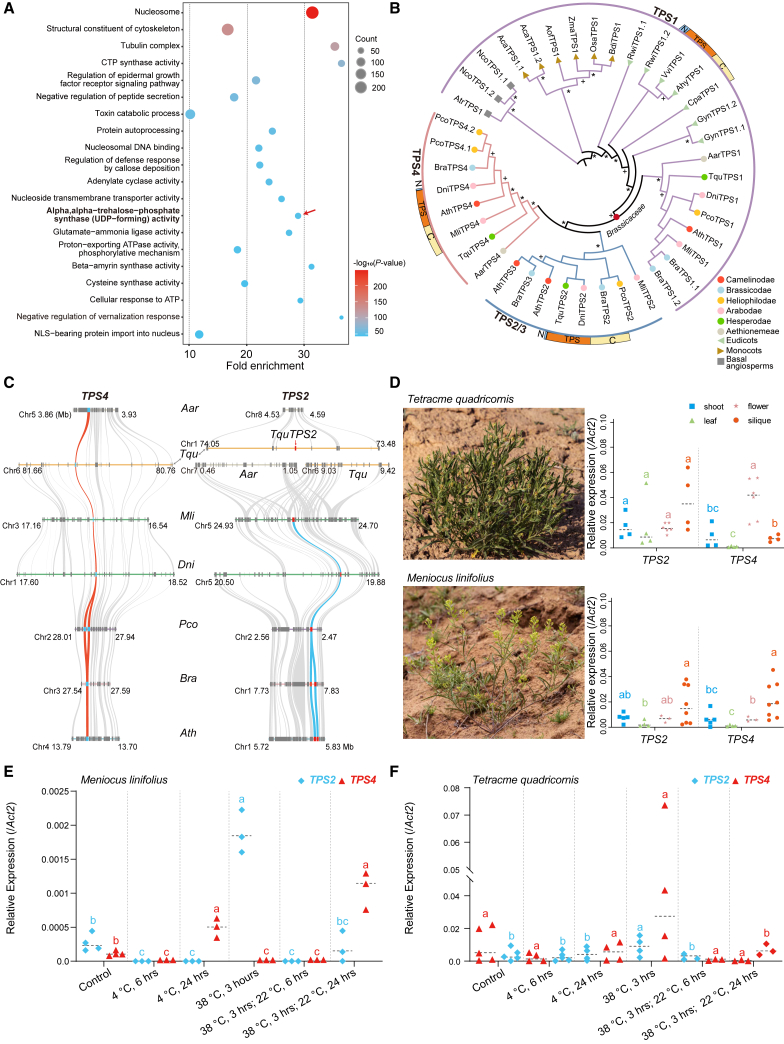
Figure 5.
Brassicaceae-specific expansion of Trehalose-6-Phosphate (T6P) synthase 1 (TPS1) genes.
(A) Gene ontology (GO) enrichment of molecular functions for expanded gene families on node F (Figure 1F) corresponding to the split between core Brassicaceae and Aethionemeae. GO terms in bold and marked with red arrows highlight the expansion of trehalose-6-phosphate (T6P) synthase-related genes.
(B) An ML tree shows the phylogenetic clustering of class I TPS proteins in 19 angiosperms, including 7 Brassicaceae species representing all supertribes and the basal Aethionemeae. The ∗ and + indicate bootstrap supports of >90% and 50%–90%, respectively. The summarized protein structure for each subgroup is shown, with N, TPS, and C indicating the N-terminal domain, TPS domain, and C-terminal domain, respectively. Supertribes or outgroups are indicated with colored circles, triangles, and squares, as shown at the bottom right.
(C) The collinearity relationships of subgroups TPS2 and TPS4 in Brassicaceae. The red and blue curves show the positions and syntenic relationships of TPS2 (red squares) and TPS4 (blue squares) between selected species. Gray curves show the surrounding syntenic genes between species.
(D) Relative expression patterns of TPS2s and TPS4s in different tissues (right panels) of plants grown under natural conditions in Fukang County, Xinjiang Province, China. A minimum of four biological replicates of leaves, shoots, flowers, and siliques were collected and pooled from at least five individuals of each species (left panels).
(E and F) Relative expression patterns of TPS2s and TPS4s under fluctuating temperature conditions for Mli(E) and Tqu(F). A minimum of three biological replicates were used for each assay. Lowercase letters indicate statistically significant differences (ANOVA, P < 0.05).