Significance
Atypical Chemokine receptor 3 (ACKR3) is a promiscuous G protein-coupled receptor (GPCR) that is intrinsically biased for arrestin signaling and an appealing drug target in numerous diseases. Here, we combined hydrogen/deuterium exchange mass spectrometry (HDX-MS), site-directed mutagenesis, and molecular dynamics (MD) simulations to identify ACKR3 conformational dynamics of activation/inhibition by various ligands. We achieved ~90% of the receptor sequence coverage in HDX-MS and milliseconds of enhanced MD sampling, which is state of the art in the GPCR field. These technical achievements allowed us to determine the inactive state of ACKR3 and its activation mechanism, identify the structural basis of ACKR3 promiscuity and hot spots for its intrinsic β-arrestin bias, and characterize the binding mode of β-arrestin 1 recruitment by ACKR3.
Keywords: HDX-MS, MD simulations, GPCR conformational dynamics, ACKR3
Abstract
Atypical Chemokine Receptor 3 (ACKR3) belongs to the G protein-coupled receptor family but it does not signal through G proteins. The structural properties that govern the functional selectivity and the conformational dynamics of ACKR3 activation are poorly understood. Here, we combined hydrogen/deuterium exchange mass spectrometry, site-directed mutagenesis, and molecular dynamics simulations to examine the binding mode and mechanism of action of ACKR3 ligands of different efficacies. Our results show that activation or inhibition of ACKR3 is governed by intracellular conformational changes of helix 6, intracellular loop 2, and helix 7, while the DRY motif becomes protected during both processes. Moreover, we identified the binding sites and the allosteric modulation of ACKR3 upon β-arrestin 1 binding. In summary, this study highlights the structure-function relationship of small ligands, the binding mode of β-arrestin 1, the activation dynamics, and the atypical dynamic features in ACKR3 that may contribute to its inability to activate G proteins.
Chemokine receptors are class A G protein-coupled receptors (GPCRs) critical for immune cell migration, immune modulation, wound healing, inflammation, and host–pathogen interactions (1). Most chemokine receptors are able to signal through the canonical G protein-mediated pathways. However, a small subset of “atypical” chemokine receptors (ACKRs) is unable to activate G proteins. ACKRs shape chemokine gradients via chemokine scavenging and dampen inflammation through β-arrestin-dependent internalization pathways (2). Among these, atypical chemokine receptor 3 (ACKR3) (formerly known as CXCR7) (3) has also been reported to bind nonchemokine ligands and scavenge opioid peptides (4). ACKR3 also interacts with various other membrane receptors (e.g., CXCR4) and alters their subcellular distribution and signaling (5). Despite possessing the general architecture and conserved sequence motifs of class A GPCRs, ACKR3 appears to exclusively activate β-arrestins (6, 7). Several cryo-EM structures of ACKR3 were recently resolved in active state, bound to its chemokine ligand CXCL12 and/or a small-molecule agonist CCX662 (8). These structures exhibit the hallmarks of canonical class A GPCR activation, mainly the outward displacement of the transmembrane helix 6 (TM6) on the intracellular side. Yet, several distinct features were observed, including the atypical orientation of the CXCL12 agonist, a kink at the C terminus of TM6–and the lack of a kink at the N terminus of TM4 into the intracellular loop 2 (ICL2). It is unclear whether these features are responsible for the atypical function of ACKR3 or are rather associated with the binding of the synthetic Fabs. In the cryo-EM structure without the intracellular Fabs, ICL1-3, and helix 8 (H8) all appeared disordered. A recent NMR study (9) highlighted that agonist-bound ACKR3 conformations differed from the antagonist-bound ones at the intracellular probe M1383.46 [superscript refers to Ballesteros and Weinstein nomenclature (10)], a known microswitch of class A GPCR activation. This reflected the TM6-TM7 movements upon ACKR3 activation, as later revealed by the cryo-EM structures. Kleist et al. also found that point mutations at N1273.35, part of the sodium-binding site, drastically affected ACKR3 activation, similar to previous findings in CXCR4 (11). Therefore, it is still unclear which structural properties distinguish ACKR3 from canonical class A GPCRs and confer its selectivity toward β-arrestin signaling. Moreover, structural features of the inactive ACKR3 state are unknown such that the molecular bases underlying the transition between inactive and active conformations are poorly understood and require further investigation.
GPCRs activation is a dynamic process during which the receptor oscillates between discrete conformational states, the most populated free energy state being in general the lowest-energy inactive state (12, 13). Ligand binding induces conformational changes in the receptor, shifting its conformational equilibrium and influencing its activity depending on the ligand efficacy. Advances in structural biology approaches have provided great insights into the diversity of GPCRs conformations (14, 15). Numerous high-resolution GPCR structures of various conformational states have revealed high diversities in ligand recognition yet striking similarities in receptor conformational changes upon activation. However, the majority of available cryo-EM and X-ray crystal structures are static snapshots of the most stable and/or accessible states under the experimental conditions used. Therefore, to understand the full complexity of GPCR conformational landscapes, complementary biophysical techniques are required to address transient states that might be discarded during sample preparation or data processing stages. Importantly, integration of several of these biophysical and computational techniques has proven especially useful for probing GPCR activation dynamics (16–20).
Hydrogen/deuterium exchange coupled with mass spectrometry (HDX-MS) is one such approach that provides a comprehensive dynamic view of GPCR structural transitions upon binding to various ligands (21–26) or intracellular partners (27, 28). HDX-MS probes the exchange between backbone amide hydrogens and deuterium present in the solvent, which is mainly related to the amide hydrogen solvent accessibility and hydrogen bond (H-bond) stability. Therefore, it is a technique that reports on the stability of protein secondary structures, the binding interface with partners, as well as the overall protein conformational dynamics (29, 30). When coupled with molecular dynamics (MD) simulations (31), HDX-MS offers the validation of the MD predictions, which together can reveal detailed receptor–ligand interactions and the conformational dynamics underlying receptor activation and inhibition.
Here, we combine HDX-MS analysis, MD simulations and site-directed mutagenesis to study the conformational changes of ACKR3 induced by a small-molecule agonist and two inverse agonists. These changes revealed important structural features associated with ACKR3 activation, including both commonalities with class A GPCRs and unique features specific to ACKR3. Namely, the agonist altered the receptor conformation at the orthosteric pocket, triggering TM6 opening on the intracellular side and high flexibility of ICL2. Inverse agonists, by contrast, locked the receptor in an inactive state and decreased the dynamics throughout the receptor. In addition, all ligands resulted in shielding of the DRY motif, which was only exposed in the presence of β-arrestin 1 in the basal state of ACKR3. Furthermore, β-arrestin 1 binding significantly reduced the deuterium uptake of the intracellular loops of ACKR3. Overall, our results provide insights into the activation mechanism of ACKR3 and its propensity toward β-arrestin recruitment.
Results
Optimization of HDX-MS to Probe ACKR3 Conformational Changes upon Ligand Binding.
We developed an HDX-MS strategy using detergent-purified ACKR3 to identify the dynamic conformational changes in ACKR3 upon ligand binding. A more native environment such as nanodiscs would provide more insights into the receptor dynamics and the role of lipids therein. However, by analyzing ligand-bound and apo receptor under identical conditions, the impact of the environment may effectively balance out, allowing for a robust comparison of the ligands’ effects. This has been previously demonstrated for the transporter BmrA that gave similar differential HDX (ΔHDX) profiles for detergent-solubilized and nanodisc-reconstituted samples (32). Three small-molecule ligands were studied, which had been characterized for their pharmacological effect toward β-arrestin recruitment: an agonist VUF15485 (33, 34) and two inverse agonists VUF16840 and VUF25550 (35–37) (Fig. 1A). All tested ligands have been shown to compete with CXCL12, an endogenous ACKR3 ligand. Extensive optimization of the receptor purification and digestion conditions (see Materials and Methods for details) allowed identification of 100 to 140 peptides covering around 90% of the receptor sequence, with an average peptide redundancy above three, after manual inspection (HDX summary tables, Dataset S1).
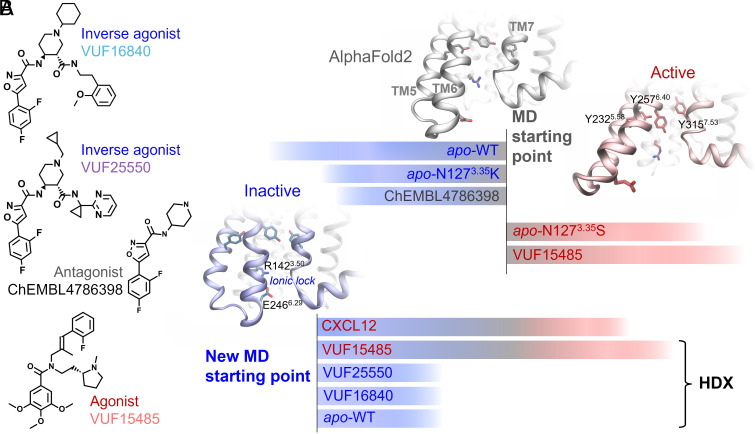
Fig. 1.
Small-molecule ligands and MD systems studied. (A) Chemical structures of the agonist VUF15485, inverse agonists VUF16840 and VUF25550, and an antagonist ChEMBL4786398. ChEMBL4786398 is a substructure of the inverse agonists, which was used here to facilitate the binding mode prediction. (B) Two batches of MD simulations from different starting structures. Initially, five simulations were performed starting from the AF model, including WT ACKR3, a constitutively active mutant (N1273.35S), an inactive mutant (N1273.35K) in apo form, as well as WT ACKR3 bound to VUF15485 and ChEMBL4786398. The agonist-bound receptor and the constitutively active mutant evolved toward active states. The other three systems evolved toward inactive states featured by TM6 inward movements and the TM6-TM3 ionic lock formation. The final inactive state of apo WT ACKR3 was then used as the new starting point to investigate the receptor conformational changes induced by agonists and inverse agonists. HDX protection factors were calculated from the new trajectories using the apo form as a reference state. These were compared with the HDX-MS data to validate the MD sampling.
In order to determine the effects of ligand binding on the ACKR3 conformations, we performed ΔHDX analysis between apo and ligand-bound ACKR3. Two to three biological replicates were performed for every condition (HDX summary tables and data tables, Datasets S1 and S2). The receptor N terminus, C terminus, and loops showed higher overall deuteration due to their unstructured properties and high flexibility (SI Appendix, Fig. S1). Conversely, we observed lower overall deuteration in the peptides corresponding to the helical transmembrane domains, reflecting their well-structured nature and low solvent accessibility due to the presence of the detergent micelle (SI Appendix, Fig. S1). Changes in these regions upon adding the ligands, if present, were thus mostly observed at longer deuteration times. We next used the ΔHDX experimental data to validate the MD simulations of ACKR3 in the presence of the three different ligands.
ACKR3 in Inactive State Conformation.
MD simulations were performed to study the conformations of ACKR3 in apo and ligand-bound forms. We used the REST2 technique (38) to enhance the MD sampling (see Materials and Methods for details). REST2 has proven very efficient in sampling conformational changes of GPCRs and other transmembrane proteins in our previous work (16, 17, 39–42). HDX protection factors (PF) were calculated from the simulation trajectories using HDXer (43), which enabled a direct qualitative comparison with the deuterium uptake data from HDX-MS. The enhanced sampling techniques were crucial for achieving the timescale necessary for an accurate alignment between MD and HDX-MS observations. This validated the MD prediction of the ligand binding poses and receptor conformational changes and gave insights into the mechanism of action of the agonist vs. inverse agonists.
The receptor conformation in apo form is a key reference for the calculation of ligand-induced HDX protection factors. We first attempted to obtain the apo conformations by REST2 MD simulations starting from the active state in the cryo-EM structures of ACKR3 (8). However, due to the short helix at ICL3 (or the TM6 kink), the receptor showed high mobility in this region and unfolding of TM6 even in the presence of the inverse agonists. This suggests that the TM6 kink is unfavorable without the intracellular Fab used for the cryo-EM structure. We therefore built an AlphaFold2 (AF) model of ACKR3, which turned out to be nearly identical to the cryo-EM structures except for the TM6-ICL3 region. It has a longer TM6 until residue K2476.30 and a shorter ICL3 without the helical kink, similar to that in the inactive structures of CXCR1, CXCR2, and CXCR4 (PDBs 2LNL, 6LFL, and 3ODU). Using the AF model as a starting point of MD simulations, we found that the apo receptor stabilized into an inactive conformation typical of class A GPCRs, in which TM6 moved significantly inward on the intracellular side (Fig. 1). To assess that the TM6-closed conformation indeed represented the inactive state, we introduced various ligands and point mutations into the AF model and performed the same simulations. The antagonist and inactive mutation also led to the TM6 inward movements (Fig. 1). By contrast, the agonist and the constitutively active mutation resulted in further openings of TM6 on the intracellular side. The agonist-bound ACKR3 also exhibited Y2325.58-Y2576.40-Y3157.53 π-stacking as TM6 opened, same as in the CXCL12-bound cryo-EM structure (Fig. 1B). In the inactive state, an ionic lock was formed between R1423.50 and E2466.29 (SI Appendix, Fig. S2A), equivalent to the conserved R3.50-X6.30 in other class A GPCRs (44). The one-residue shift in ACKR3 ionic lock compared to the conserved lock at the level of TM6 may weaken the ionic interaction, which could contribute to the high basal activity of this atypical receptor (45). To test this hypothesis, we performed site-directed mutagenesis and found that interrupting the ionic lock increased both the basal activity and the agonist-dependent recruitment of β-arrestin 1 to ACKR3 (SI Appendix, Fig. S2B). The E2466.29K mutation showed stronger effects than E2466.29A, while all mutants maintained the affinity for the agonist comparable to that of the WT (SI Appendix, Fig. S2B). These results suggest that ACKR3 conserves the ionic lock of class A GPCRs and the common conformational changes upon activation, whereas the initial AF model may represent an intermediate or “active-like” state. Therefore, we chose the apo ACKR3 model in inactive state as the reference apo state and a new starting point of MD simulations, to study the impact of different ligands on the receptor conformation. The MD simulations of ACKR3 bound to ChEMBL4786398 served to deduce the binding mode of the more challenging analogs, VUF16840 and VUF25550 (Fig. 1A). We did not perform HDX-MS with ChEMBL4786398 which is less potent than the two inverse agonists of interest. The MD-predicted ligand binding mode and receptor dynamics were in good agreement with the HDX-MS and site-directed mutagenesis data, as discussed below. Additional simulations were performed using reservoir replica-exchange MD (R-REMD) (46, 47) to further validate the findings (Materials and Methods and SI Appendix, Supplementary Methods).
Ligand Binding Mode and Protection at ACKR3 Orthosteric Site.
The HDX-MS data showed a decreased deuterium uptake at the extracellular face of ACKR3 in the presence of all tested ligands, regardless of their pharmacological profile (Fig. 2 and SI Appendix, Fig. S3). Namely, we observed protection to deuteration at the level of the receptor orthosteric binding pocket. The most significant protection was at several peptides spanning the extracellular top of TM5. In the case of peptide 214 to 220 at TM5, protection was up to 35% and 24% by the two inverse agonists and 17% by the agonist. The top of TM4 was also highly protected, mainly with the inverse agonists (Fig. 2). For peptide 175 to 181 at TM4 for instance, inverse agonists VUF25550 and VUF16840 induced a decrease in deuteration whereas the agonist VUF15485 induced a slight protection at longer deuteration times. Of note, significant deprotection was observed for peptide 175 to 181 at short deuteration timepoints in response to the agonist (Fig. 2B; TM4); however, this was not reproducible. This highlights the importance of performing biological replicates when performing HDX-MS analysis.
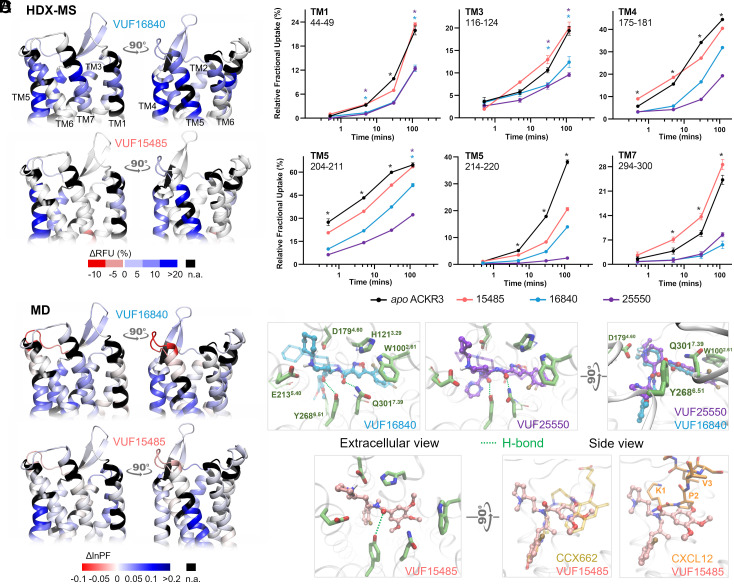
Fig. 2.
Ligand binding at the level of ACKR3 orthosteric pocket. (A) Schematic representation of the % differential relative fractional uptake data (apo ACKR3—bound ACKR3) mapped onto the upper part of the AF model of ACKR3 (for clarity, we did not include the N terminus). Relative fractional uptake is calculated by dividing the experimental uptake (Da) of a peptide by its maximum possible uptake. This depicts reproducible and statistically significant ΔHDX in response to inverse agonists small ligands represented by VUF16840 (VUF25550 giving a similar profile), or agonist small ligand VUF15485. Black regions represent regions with no sequence coverage. Ligand-induced reduction in deuterium uptake is represented in blue while ligand-induced increase in deuterium uptake is in red, according to the scale. (B) Associated deuterium uptake plots showing the relative uptake for peptides from apo or ligand-bound ACKR3 across several deuteration time points and are representative of the extracellular region as indicated at the Top of the plot. Statistically significant changes were determined using Deuteros 2.0 software (48) (P ≤ 0.01): statistically significant time points for the different ligands are represented by a colored star corresponding to the ligand in question (pink, blue, and purple for VUF15485, VUF16840, and VUF25550 respectively). Black stars depict time points that are statistically significant for all three ligands. Uptake plots are the average and SD of three technical replicates from the same biological preparation of ACKR3. (C) Calculated differential ln HDX protection factor changes (ΔlnPF) mapped on the AF model. Per-residue lnPF was first calculated for each MD trajectory of ACKR3 in apo and ligand-bound forms. The difference between the apo and bound forms gave the per-residue ΔlnPF for each ligand. For comparison with the HDX data, per-peptide ΔlnPF was calculated by averaging the per-residue ΔlnPF over the peptides obtained in the HDX-MS experiments for each ligand (see Materials and Methods and Dataset S3 for details). (D) Predicted ligand binding mode. The inverse agonists could bind in both enantiomers of the piperidine group (solid and transparent depictions). They showed nearly identical binding poses except that the cyclopropyl-pyrimidine of VUF25550 was more mobile. The agonist VUF15485 formed ionic interactions with D1794.60 via its 1-methylpyrrolidine. The rest of VUF15485 largely overlaps with CCX662 as shown in the superimposition to PDB 7SK8. It also overlaps with the N terminus of CXCL12.
The MD prediction of the ligand binding mode and PF were coherent with the HDX-MS data (Fig. 2C). Discrepancies were mostly in the loop regions, near the truncated N- and C- termini, and in regions where HDX-MS showed no significant changes. All the three small-molecule ligands anchored to D1794.60 via the amine (Fig. 2D), resembling the role of residue K1 in CXCL12 (8). The inverse agonists, VUF16840 and VUF25550, could bind in both enantiomers of the piperidine, which anchored to D1794.60 and E2135.40 through electrostatic interactions, respectively. The agonist VUF15485, however, was only stable in one enantiomer in the simulations, which anchored to D1794.60. VUF15485 occupied the pocket space that largely overlaps with the partial agonist CCX662 and the N terminus of CXCL12 in the cryo-EM structures (PDBs 7SK8 and 7SK3). It formed a H-bond with Y2686.51 via its amide carbonyl. The two inverse agonists extended further in the pocket toward TM2. They showed more persistent interactions than the agonist, namely π–π stackings with W1002.61 and H1213.29, and an additional H-bond with Q3017.39. These explain the stronger protections in TM2, TM3, and TM7 in HDX-MS. The predicted ligand binding poses are confirmed by site-directed mutagenesis data on VUF15485 and VUF16840 binding (SI Appendix, Fig. S4 and Table S1). Namely, a D1794.60N mutation diminished the binding affinities for both ligands, whereas E2135.40Q only affected VUF16840. In addition, Q3017.39E/A mutations diminished the affinity for VUF16840 but not VUF15485. A D2756.58N mutation had no impact on either ligand, since the mutation site lies on the extracellular rim of the orthosteric pocket, distant from the predicted ligand-binding site. Eight other mutations in the orthosteric pocket were tested to validate the binding pose of VUF15485 (SI Appendix, Fig. S4 and Table S1) (34). Mutations W100Q2.61 and F124A3.32 and Y268A6.51 abolished the ligand binding, since all the three aromatic residues formed primary pi-stacking and hydrophobic contacts with the ligand. Y2686.51 also engaged in hydrogen bonding. Mutation of other binding-site residues showing no specific interactions with VUF15485 had negligible impact on the ligand binding.
The HDX-MS data at the orthosteric pocket of ACKR3 and the MD predicted ligand binding mode are consistent with the ability for all tested ligands to inhibit CXCL12 binding. Cryo-EM structures of ACKR3 in complex with CXCL12 show that CXCL12-K1 and P2, as part of the receptor activation motif of CXCL12, penetrate ACKR3 orthosteric pocket to make side chain interactions with E2135.40, D1794.60, and Y200ECL2. CXCL12-V3 backbone amide was also shown to potentially form a H-bond with ACKR3-Q3017.39 in addition to packing of V3 side chains against proximal residues, L2977.35 and H2987.36. MD predictions show that the small-molecule ligands overlap largely with CXCL12-K1 and P2 (Fig. 2D). In correlation, HDX uptake is reduced for peptides spanning these residues where all the ligands show main contacts at the level of TM4 and TM5. Protective effects on TM7 were mainly visible for the inverse agonists which form two additional H-bonds with Y2686.51 and Q3017.39 (Fig. 2). Taken together, our data indicate overlapping contacts for CXCL12 and the small-molecule ligands at the orthosteric binding pocket.
All Ligands Shield the DRY Motif.
ΔHDX-MS showed that binding of the different ligands led to significant allosteric changes in the intracellular side of ACKR3. Notably, the DRY motif at the intracellular face of TM3 was protected in the presence of all ligands, implying that this motif is always shielded upon binding of these ligands (Fig. 3). The agonist-induced protection on the representative peptide 138 to 143 spanning the DRY motif was weaker than the inverse agonists (Fig. 3 and SI Appendix, Fig. S5). MD simulations suggest that in the inverse agonist-bound states, the DRY is shielded by TM6 and protected to deuterium uptake by the overall decrease of receptor conformational dynamics, whereas in the agonist-bound states, the DRY is shielded by both TM5 and Y2576.40 (SI Appendix, Fig. S5). The Y2576.40 orientation here is similar to the one observed in the CXCL12-bound cryo-EM structure. This agrees with the changes in the NMR spectra of M1383.46 upon agonist binding, where an aromatic residue was detected in its proximity (9). Indeed, we observed in the MD simulations that Y2576.40 was dynamic in the apo form, whereas agonists led to proximation of Y2576.40 to M1383.46 (SI Appendix, Fig. S5A). However, we did not observe a clear correlation between the M1383.46 side-chain conformations and the ligand efficacies (SI Appendix, Fig. S5B). The different Y2576.40 dynamics in the apo and agonist-bound states may explain the fact that Y2576.40 is critical for the ACKR3 basal activity but not the agonist-dependent arrestin recruitment (8).
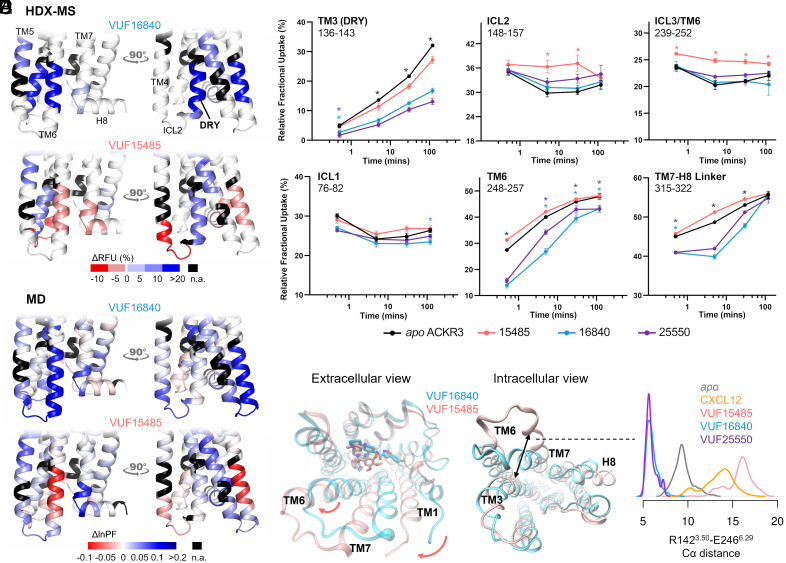
Fig. 3.
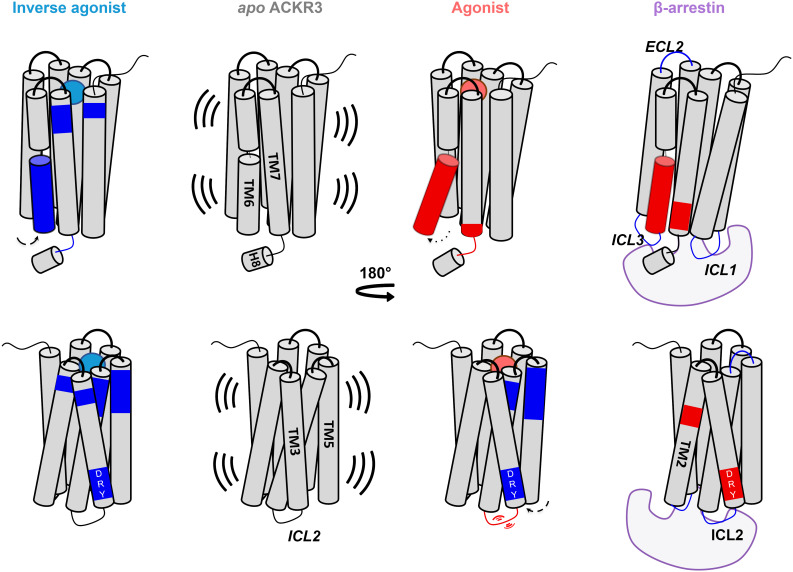
Allosteric conformational changes of ACKR3 activation. (A) Schematic representation of the % differential relative fractional uptake data (apo ACKR3—bound ACKR3) mapped onto the lower part of the AF model of ACKR3 (for clarity, we did not include the C- terminus). Relative fractional uptake is calculated by dividing the experimental uptake (Da) of a peptide by its maximum possible uptake. Scheme depicts reproducible and statistically significant ΔHDX in response to inverse agonists small ligands represented by VUF16840 (VUF25550 giving a similar profile), or agonist small ligand VUF15485. Black regions represent regions with no sequence coverage. Ligand-induced reduction in deuterium uptake is represented in blue while ligand-induced increase in deuterium uptake is in red, according to the scale. (B) Associated deuterium uptake plots showing the relative uptake for peptides from apo or ligand-bound ACKR3 across several deuteration time points and are representative of the intracellular region as indicated at the Top of the plot. Statistically significant changes were determined using Deuteros 2.0 software (48) (P ≤ 0.01): statistically significant time points for the different ligands are represented by a colored star corresponding to the ligand in question (pink, blue, and purple for VUF15485, VUF16840, and VUF25550 respectively). Black stars depict time points that are statistically significant for all three ligands. Uptake plots are the average and SD of 3 technical replicates from the same biological preparation of ACKR3. (C) Calculated ΔlnPF from the MD simulations, mapped on the AF model. (D) Proposed mechanism of activation. The bulky trimethoxybenzamide group of VUF15485 induces a “twist” of the 7TM bundle around the orthosteric pocket, which allosterically triggers TM6 opening on the intracellular side. (E) Probability density distribution of the ionic-lock-residue Cα distances during the MD simulations.
The DRY motif is highly conserved across GPCRs and has been shown to be play a role in G protein binding and in maintenance of the ionic lock. Particularly, R1423.50 has been shown to have direct G protein contacts in structures of several GPCR-G protein ternary complexes (49). In some cases, R1423.50 was also shown to have direct contacts with β-arrestin (50, 51). Since all the ligands we tested induced a protection at this region, this could be related to the inability for ACKR3 to activate G proteins.
Typical and Distinct Features of ACKR3 Activation.
The MD simulations of ACKR3 bound to CXCL12 and VUF15485 both displayed a remarkable TM6 opening on the intracellular side (Fig. 3 D and E), typical of class A GPCR activation. The intracellular half of TM6 was deprotected, as shown by the lower lnPF than the apo form calculated from the MD trajectories (Fig. 3C). This was confirmed by the HDX-MS results, in which the lower half of TM6 showed higher uptake in the presence of VUF15485 compared to the apo receptor (Fig. 3 A and B). Interestingly, ΔHDX showed that I2546.37 was protected with inverse agonists and deprotected with the agonist, which correlates with the observed interactions between I2546.37 and M1383.46 only in inactive structures of GPCRs (9). The TM7-H8 linker, however, showed discrepancies between the MD and the HDX-MS data in the presence of the agonist. This may be due to the C-terminal truncation in the MD and/or to insufficient sampling of the apo form, where the inactive state was overstabilized by the ionic lock (Fig. 3E). Although ACKR3 has a high basal activity, the apo state did not show clear activation during the MD simulations, which likely overestimated the ΔlnPF of the agonist-bound state. The high basal activity of ACKR3 is reflected by the less pronounced absolute ΔHDX values upon agonist-binding compared to inverse agonist-binding. This also correlates to a recent single-molecule Förster resonance energy transfer (FRET) study (52) demonstrating substantial populations of active and intermediate states of apo ACKR3. ICL2 and the intracellular tip of TM4 also exhibited significant deprotection upon agonist binding, in both the MD simulations and the HDX-MS experiments (Fig. 3). The MD simulations indicate that VUF15485 led to high ICL2 mobility and unfolding of the TM4 intracellular tip upon TM6 opening. Nevertheless, it is worth noting that loop conformations are often undersampled in MD simulations. Interestingly, ICL2 in most class A GPCRs forms a short α-helix in both inactive and active states. ICL2 has been associated with G protein subtype selectivity and G protein signaling of several receptors (27, 28, 53). Further investigation is required to understand the potential role of ACKR3 ICL2 in the lack of G protein signaling of this atypical receptor.
Taken together, our data suggest the following mechanism of activation by the agonist: VUF15485 may induce an overall twist of the 7TM bundle, due to the bulky trimethoxy-benzamide ring in the center of the orthosteric pocket (Fig. 3D). The twist propagates to the intracellular side allosterically in a loosely coupled manner, which triggers TM6 outward movements. A similar phenomenon was observed in the simulations of CXCL12-bound ACKR3 (SI Appendix, Fig. S6). The inverse agonists, by contrast, may stabilize the inactive state of the 7TM bundle through additional interactions with the receptor within the orthosteric pocket. Namely, the inverse agonists, which are longer but less bulky, extend toward TM2 to TM3. They form additional interactions with TM2, TM3 (π-stacking with W1002.61, H1213.29), and TM7 (H-bond with Q3017.39), which may restrain receptor conformational changes (Fig. 2D). Consistently, the inverse agonists resulted in additional protection on TM3 and TM7 in the orthosteric pocket, as well as overall protection of the receptor from HDX (Fig. 2). Mutation Q3017.39A has been shown to significantly increase the ACKR3 basal activity (8). Here, our findings indicate that Q3017.39 is also key for the ligand-dependent activity. Indeed, ligand-TM7 contacts have been reported to be important for β-arrestin signaling bias in other class A GPCRs (9, 19, 54–56). Although there appears to be no common pattern across different GPCR families, the pocket area between TM2 and TM7 is likely a hot spot for ligand bias. The above data show that agonist-induced ACKR3 activation increases the solvent accessibility of TM6, ICL2, TM7, and the TM7-H8 linker on the intracellular side, suggesting that these conformational changes occur to facilitate the recruitment of β-arrestins.
HDX-MS Identifies Putative β-Arrestin Binding Sites.
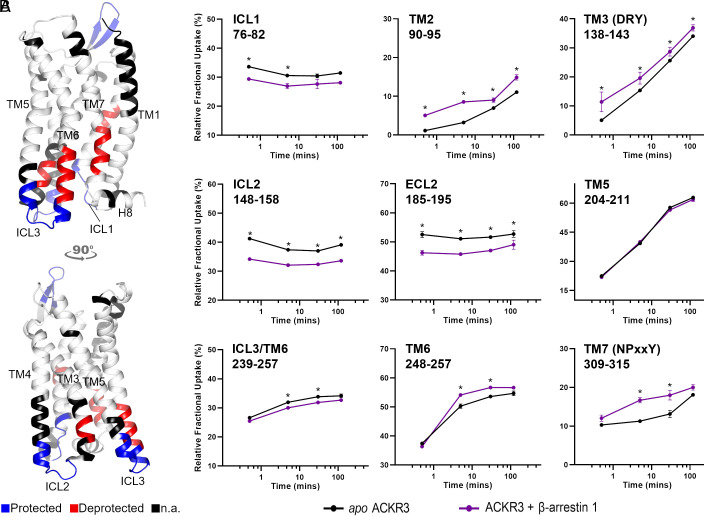
To follow-up on ACKR3 binding to β-arrestin 1, we took advantage of the constitutive activity of the receptor and performed ΔHDX analysis of ACKR3 in the presence of 1.2 molar equivalent β-arrestin 1 ΔCter (Fig. 4). As the C-tail of β-arrestin has an autoinhibitory function, its removal was necessary to shift β-arrestin 1 toward the active state (57). The presence of β-arrestin 1 led to the decreased deuteration of ICL2 and ICL1 and, to a lesser extent, of ICL3 (Fig. 4). Interestingly, peptides spanning the NPxxY motif as well as the intracellular face of TM6 showed increased deuteration upon β-arrestin 1 binding. Another notable finding was the deprotection of the DRY motif in the presence of β-arrestin 1 (representative peptide 138 to 143, Fig. 4). Therefore, the ACKR3 DRY motif becomes more exposed and/or dynamic upon binding to β-arrestin 1, suggesting that this motif is not implicated in the direct interaction with β-arrestin 1. Deprotection was also visible at the level of mid-TM2 (representative peptide 90 to 95), a region adjacent to the highly conserved D902.50 which is part of the sodium binding site in class A GPCRs. Sodium ions are known negative allosteric modulators of class A GPCR activity (58). The deprotection observed at this level implies allosteric cooperation between the ligand/sodium-binding sites and β-arrestin binding, which may be explored for ligand design. Finally, we observed some allosteric effects upon ACKR3 binding to β-arrestin 1, the most notable one being the ECL2 protection (Fig. 4).
Fig. 4.
β-Arrestin 1 binding to ACKR3. (A) Schematic representation of the protected (blue) and deprotected (red) regions of ACKR1 in the presence of β-arrestin 1 mapped onto the AF model of ACKR3 (for clarity, we did not include the N- and C- termini). This depicts reproducible and statistically significant changes in deuterium uptake in the presence of β-arrestin 1. Black regions represent regions with no sequence coverage. (B) Associated deuterium uptake plots showing the relative uptake for peptides from apo or arrestin-bound ACKR3 across several deuteration time points and are representative of the region as indicated at the Top of the plot. Statistically significant changes are represented by black stars and were determined using Deuteros 2.0 software (48) (P ≤ 0.01). Uptake plots are the average and SD of three technical replicates from the same biological preparation of ACKR3.
The overall ΔHDX profile suggests that engagement of β-arrestin 1 at ACKR3 involves all ICLs, possibly in a similar fashion to those reported for the V2R (59) and the neurotensin receptor type 1 (NTSR1) (60), which both engage β-arrestin with all three ICLs. The 10 top-ranked AF models of ACKR3 and β-arrestin 1 complex we generated (SI Appendix, Fig. S7) all showed that the middle loop and finger loop of β-arrestin interact with the receptor intracellular pocket, ICL1, 2, and 3. The multiple interactions observed between ACKR3 ICLs and β-arrestin 1 correlate with the observed decrease in deuterium uptake for these loops in the presence of β-arrestin 1. Often ICL3 is not well resolved in high-resolution GPCR structures. However, ICL1 and ICL2 have been highlighted as important interfaces for β-arrestin binding. In the majority of published cryo-EM complexes, ICL2 sits in a defined cleft between the N and C domains of β-arrestin (50, 51, 59, 61). The only known exception is NTSR1 where ICL1 takes this position instead (62). Additionally, ICL1 has been shown to contribute to β-arrestin binding for all GPCR/β-arrestin complexes apart from that of the M2R (61). The increased deuterium uptake observed at ICL2 in response to the agonist and the protection of all ICLs in the presence of β-arrestin 1 therefore suggests that these regions mediate the interaction with β-arrestin. To evaluate this role of ICL2, we tested a chimeric ACKR3 in which residues 144ICL2 to 149ICL2 of ACKR3 were replaced with corresponding residues of CXCR4. Maximum β-arrestin 1 recruitment was significantly reduced for this ICL2 chimera despite it having a comparable affinity to the WT for CXCL12 (SI Appendix, Fig. S7D). These data, together with our findings that orthosteric ligands affect ICL2 allosterically, indicate a major role of ICL2 in β-arrestin 1 binding to ACKR3.
Discussion
Being an intrinsically β-arrestin-biased chemokine receptor, ACKR3 is an interesting subject for the study of GPCR functional selectivity. Its promiscuity for nonchemokine ligands and selectivity for chemokines add to its intriguing character. It is necessary to understand this functional bias in order to fully elucidate the role ACKR3 plays in tumor microenvironments where it is often overexpressed (63, 64) or to maximize the cardioprotective effects associated with ACKR3 (65). Combining HDX-MS and MD analysis, we identified the inactive state of ACKR3, as well as typical and atypical conformational features of ACKR3 activation (Fig. 5). It has been suggested that ACKR3 may have an intrinsically active-like structure (8), given its high basal activity. Our data show that ACKR3 adopts typical class A GPCR inactive-active conformational changes, featuring the conserved TM3 to TM6 ionic lock in inactive state and TM6 opening upon activation. This is triggered by an overall twist of the 7TM bundle upon agonist binding, which propagates allosterically to the intracellular side in a loosely coupled manner. The inverse agonists, by contrast, stabilized the inactive state of the 7TM bundle through additional interactions in a subpocket between TM2, TM3, and TM7. β-arrestin 1 binding led to HDX deprotection of TM2 immediately below the TM2-TM7 subpocket, which has previously been reported as a hot spot for GPCR ligand bias (19, 56).
Fig. 5.
Scheme summarizing changes induced by binding of an agonist, inverse agonist and β-arrestin 1 to ACKR3. Activation through agonist binding induces an allosteric opening of TM6 and resulted in increased flexibility and/or solvent exposure (red regions) of peptides spanning TM6, TM7, and ICL2. Inverse agonists resulted in decreased flexibility and/or solvent exposure (blue regions) through receptor constraint. β-arrestin 1 binding reduced the deuteration of all intracellular loops and resulted in increased deuteration of the NPxxY motif (TM7), DRY motif (TM3), and the conserved sodium binding site D902.50 (TM2). Arrows depict movements for the respective helices in response to different binders. Activation is associated with outward movement of TM6 and inward movement of TM5 which contributes to protection of the DRY motif in the presence of the agonist.
Agonist binding also led to significant allosteric deprotection of ICL2 and the intracellular tip of TM4, correlating to an increased flexibility in this region. MD simulations indicated that as TM6 moved outward, the contacts between TM5-ICL3-TM6 and TM3 changed remarkably, leading to deprotection of this region. Mutating ICL2 significantly reduced β-arrestin 1 recruitment (discussed below), indicating the importance of this loop for β-arrestin 1 binding. It is yet to be investigated whether ICL2 plays a role in the lack of G protein signaling of ACKR3.
Another intriguing observation is that the DRY motif was protected upon ligand binding, even after opening of TM6 in the presence of the agonist. It appeared to be shielded by Y2325.58, Y2576.40, and Y3157.53, which formed π-stacking in the active state. Y2576.40 is found only in ACKR3 and GPR182 among nonolfactory class A GPCRs (66). However, it is unclear whether the reduced accessibility of the DRY in ACKR3 contributes to its lack of G protein signaling. In the presence of β-arrestin 1, on the other hand, the DRY motif was slightly deprotected, suggesting that this motif is not shielded upon ACKR3 binding to β-arrestin 1. This is note-worthy as prior studies have shown that ACKR3 has the potential to interact with G-proteins (67) and even activate Gαi in astrocytes (68). β-arrestins are known to serve as scaffolds and allosteric regulators of downstream GPCR signaling pathways (69) and have been shown to participate in endosomal mega-complexes with GPCRs and G-proteins (70). Therefore, the observed deprotection at the DRY in the presence of β-arrestin 1 hints that this may facilitate association or activation of G-proteins by ACKR3. If such is the case, further understanding of this system and manipulation of ACKR3 endosomal signaling may contribute to our understanding of its role in various pathologies.
A recent preprint described cryo-EM structures of β-arrestins in complex with GRK2/5 differentially phosphorylated ACKR3 in the presence of CXCL12 and a Fab that stabilizes active β-arrestins (71). Interestingly, the authors observed unconventional interactions between β-arrestins and ACKR3. Unlike all other reported GPCR–arrestin complexes, the β-arrestin finger loop does not insert into the transmembrane core of ACKR3 and instead embeds the detergent micelle. The authors highlight that GRK2- or GRK5-phosphorylation drives ACKR3 to bind β-arrestins in discrete modes through primarily its C-tail or TM1/TM7, respectively. Our study shows that in the constitutive recruitment of β-arrestin 1 by ACKR3 and in the absence of any kinase or Fab, all ICLs of the receptor were unambiguously protected from deuterium exchange highlighting their importance in binding of β-arrestin 1. Notably, mutation of ICL2 significantly reduces β-arrestin 1 recruitment to ACKR3 in living cells (SI Appendix, Fig S7), confirming the essential role of this loop in β-arrestin 1 recruitment. Since β-arrestin binding is a highly dynamic and heterogenous process that depends on the nature of the agonist, the phosphorylation patterns, and the presence of other potential binding partners, it is not surprising that several binding patterns could be observed in vitro. Our study also shows that peptides spanning the NPxxY motif and the intracellular face of TM6 were deprotected upon β-arrestin 1 binding. Several studies have reported important roles of TM7 and the NPxxY motif in class A GPCR signaling bias (19, 72–74). Particularly, Y3157.53 is highly conserved and the Y3157.53A mutation almost completely abolishes β-arrestin recruitment by ACKR3 (9). Other studies have suggested that a twist in this region, above P7.50 can also dictate whether GPCRs are preferentially coupling to G-proteins or arrestins (75, 76). The deprotection at the NPxxY motif therefore suggests that this region becomes more exposed or more flexible in the presence of β-arrestin. Further studies are required to better understand the functional significance of this conformational change.
Taken together, these differential effects observed in response to ACKR3 activation and inverse agonism give insights into specific receptor–ligand interactions and receptor microswitches controlling ACKR3 function and bias toward β-arrestin. By synergistically integrating MD simulations and experimental HDX-MS data, we could obtain atomistic-level comprehension of receptor dynamics. These insights and tools are applicable across various drug target proteins and could help designing drugs that can elicit more specific and targeted effects, which will ultimately aid the development of new and efficacious therapeutics.
Materials and Methods
Expression and Purification of ACKR3.
For production in insect cells to obtain adequate yields for HDX-MS experiments, the full-length gene of human ACKR3 was subcloned into pFastBac1 to enable infection of sf9 insect cells. The construct bore a hemagglutinin signal peptide followed by a Flag-tag preceding the receptor sequence. ACKR3 N13, N22, and N33 residues were substituted with a Glutamine in order to avoid N-glycosylation, which contribute to sample heterogeneity and therefore hinders detection of peptides in this region. The aforementioned mutations have previously been shown to have no significant impact on chemokine binding and signaling through ACKR3 (77).
Flag-ACKR3 was expressed in sf9 insect cells using the pFastBac baculovirus system (Thermo Fisher Scientific). Cells were grown in suspension in EX-CELL 420 medium (Sigma-Aldrich) and infected at a density of 4 × 106 cells/mL with the recombinant baculovirus. Flasks were shaken for 48 h at 28 °C, subsequently harvested by centrifugation (3,000 g, 20 min) and stored at −80 °C until usage. ACKR3 was purified in the presence of ACKR3 agonist VUF11207 (Bio-Techne) to stabilize the receptor. Cell pellets were first thawed and lysed by osmotic shock in a buffer containing 10 mM Tris (pH 7.5), 1 mM Ethylenediaminetetraacetic acid (EDTA), 2 mg/mL iodoacetamide, 1 μM VUF11207, and protease inhibitors: 50 μg/mL Leupeptin (Euromedex), 0.1 mg/mL Benzamidine (Sigma-Aldrich), and 0.1 mg/mL Phenylmethylsulfonyl fluoride (PMSF; Euromedex). Lysed cells were centrifuged for (38,400 g, 10 min) and the resulting pellet was solubilized and dounce-homogenized 20× in buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 2 mg/mL iodoacetamide, 1 μM VUF11207, 0.5% (w/v) Dodecyl-β-D-maltoside (DDM) (Anatrace), Cholesteryl hemisuccinate 0.1% (w/v), and protease inhibitors (50 μg/mL Leupeptin, 0.1 mg/mL Bensamidine, and 0.1 mg/mL PMSF). The homogenate was subsequently stirred for 1 h at 4 °C and centrifuged (38,400 g, 30 min). The supernatant was then loaded onto M2 anti-Flag affinity resin (Sigma-Aldrich) using gravity flow. Resin was subsequently washed with 10 column volumes of DDM wash buffer containing 50 mM Tris, 150 mM NaCl, 0.1 μM VUF11207, DDM 0.1% (w/v), Cholesteryl hemisuccinate 0.02% (w/v). Detergent was then gradually exchanged from DDM to LMNG (Anatrace) using increasing ratios of DDM wash buffer and buffer containing 50 mM Tris, 150 mM NaCl, 0.02 μM VUF11207, 0.2% MNG (w/v), 0.05% CHS (w/v). Once detergent was fully exchanged, MNG and CHS concentrations were steadily reduced to 0.005% and 0.001% respectively. ACKR3 was finally eluted in 50 mM Tris, 150 mM NaCl, 0.02 μM VUF11207, 0.005% MNG (w/v), 0.001% CHS (w/v), and 0.4 mg/mL Flag peptide (Covalab). The eluate was concentrated using a 50 kDa molecular weight cutoff (MWCO) concentrator (Millipore), then ACKR3 was purified by size exclusion chromatography (SEC) using a Superdex 200 Increase (10/300 GL column) connected to an ÄKTA purifier system (GE Healthcare) and eluted in elution buffer [50 mM Tris, 150 mM NaCl, 0.005% MNG (w/v), 0.001% CHS (w/v)]. Final elution buffer is free from VUF11207 in order to ensure an apo state devoid of ligand, which was confirmed by native MS analysis. Fractions containing monomeric ACKR3 were concentrated to between 20 and 25 μM, aliquoted, flash-frozen, and stored at −80 °C prior to HDX experiments.
Expression and Purification of β-Arrestin 1.
A truncated form of β-arrestin1 was used in HDX experiments due to the higher basal activity of the protein. This construct was truncated at residue 382, lacking the last 36 amino acids, and produced as follows. Competent BL21 Escherichia coli cells (Thermo Fisher Scientific) were transformed using a pET plasmid containing β-arrestin1 ΔC-ter containing a Twin-Strep tag at the N terminus. Cells were cultured in LB with Kanamycin (37 °C, 170 rpm) until an optical density of 0.6 U was reached after which 0.025 mM Isopropyl-β-D-thiogalactopyranoside was added to induce cell expression. Cells were further incubated for 5 h at 37 °C and harvested by centrifugation (3,000 g). Pellets were stored at −80 °C prior to use. Protein purification was as follows. Cell pellets were thawed and resuspended in buffer containing Tris-HCl (pH8), 1 mM EDTA, 200 mM NaCl, and 1 mM β-mercaptoethanol and protease inhibitors Leupeptin (5 μg/mL), benzamidine (10 μg/mL), and PMSF (10 μg/mL). Cells were then lysed by sonication and then MgCl2 and benzonase were added to the lysate, 5 mM, and 2,000 U respectively. Lysate was centrifuged (38,400 g, 20 min, 4 °C) and supernatant was supplemented with BioLock (0.75 mL/L) and loaded onto StrepTactin affinity resin at 4 °C. The resin was then washed with a wash buffer containing 20 mM Tris (pH8), 200 mM NaCl, and 100 μM Tris(2-carboxyethyl)phosphine (TCEP). Wash buffer supplemented with 2.5 mM desthiobiotin (IBA) was used to elute the protein. The eluate was then further purified by SEC using a Superdex 200 Increase (10/300 GL column) connected to an ÄKTA purifier system (GE Healthcare) in a buffer containing 20 mM HEPES (pH 7.5), 200 mM NaCl, and 100 μM TCEP. Eluted fractions containing purified β-arrestin were collected and concentrated using a 10 kDa MWCO concentrator (Millipore). Aliquots were flash-frozen and kept at −80 °C.
HDX-MS Experiments.
HDX-MS experiments were performed using a Synapt G2-Si HDMS coupled to nanoAQUITY UPLC with HDX Automation technology (Waters Corporation). ACKR3 in LMNG detergent was concentrated up to 20 to 25 µM and optimization of the sequence coverage was performed on undeuterated controls. Various quench times and conditions were tested, i.e., in the presence or absence of different denaturing or reducing reagents with or without longer trapping times to wash them out. Different digestion enzymes were tested (Pepsin, Nepenthesin-2, Rhizopuspepsin). The best sequence coverage and redundancy for ACKR3 were systematically obtained with Nepenthesin-2 (Affipro) without the addition of any denaturing agents in the quench buffer. Mixtures of receptor: ligands were preincubated together to reach equilibrium prior to HDX-MS analysis. Analysis of freshly prepared ACKR3 apo, ACKR3: small ligands (1:10 ratio), and ACKR3: β-arrestin (1:1.2 ratio) mixtures were performed as follows: 3 µL of sample are diluted in 57 µL of undeuterated for the reference or deuterated last wash SEC buffer. The final percentage of deuterium in the deuterated buffer was 95%. Deuteration was performed at 20 °C for 0.5, 5, 30, and 120 min. Next, 50 µL of reaction sample were quenched in 50 µL of quench buffer (KH2PO4 50 mM, K2HPO4 50 mM, 200 mM TCEP pH 2.3) at 0 °C. 80 µL of quenched sample were loaded onto a 50 µL loop and injected on a Nepenthesin-2 column (Affipro) maintained at 15 °C, with 0.2% formic acid at a flowrate of 100 µL/min. The peptides were then trapped at 0 °C on a Vanguard column (ACQUITY UPLC BEH C18 VanGuard Precolumn, 130 Å, 1.7 µm, 2.1 mm × 5 mm, Waters) for 3 min, before being loaded at 40 µL/min onto an Acquity UPLC column (ACQUITY UPLC BEH C18 Column, 1.7 µm, 1 mm × 100 mm, Waters) kept at 0 °C. Peptides were subsequently eluted with a linear gradient (0.2% formic acid in acetonitrile solvent at 5 up to 35% during the first 6 min, then up to 40% and 95% over 1 min each) and ionized directly by electrospray on a Synapt G2-Si mass spectrometer (Waters). HDMSE data were obtained by 20 to 30 V trap collision energy ramp. Lock mass accuracy correction was made using a mixture of leucine enkephalin and GFP. For every tested condition we analyzed two to three biological replicates, representing different protein productions in insect cells, and deuteration timepoints were performed in triplicates for each condition.
Peptide identification was performed from undeuterated data using ProteinLynx global Server (version 3.0.3, Waters). Peptides were filtered by DynamX (version 3.0, Waters) using the following parameters: minimum intensity of 1,000, minimum product per amino acid of 0.2, maximum error for threshold of 10 ppm, and presence of peptides in at least 3 out of 6 files. All peptides were manually checked, and data were curated using DynamX. Back exchange was not corrected since we are measuring ΔHDX and not absolute one. Statistical analysis of all ΔHDX data was performed using Deuteros 2.0 (48) and only peptides with a 99% CI were considered.
AlphaFold Predictions.
A SBGrid consortium installation of AlphaFold version 2.3 (78) was run on a local server equipped with a NVIDIA Tesla P100 GPU in order to predict the structure of the ACKR3 monomer and ACKR3/β-arrestin1 heterodimer. The full databases were used, with max_template_date = 2023-04-09. The model_preset option was set to “monomer” for ACKR3 and to “multimer” for the heterodimer. All other parameters were left to their default values.
MD Simulations.
Ligands were docked into ACKR3 models using Autodock Vina. Four isomers per ligand were tested, including two diastereoisomers and two amine enantiomers. Pocket residues and ligand rotatable bonds were set as flexible. Top-ranked binding poses underwent MD simulations until a stable binding pose was obtained. The replica exchange with solute scaling (REST2) (38) technique was used to enhance the conformational sampling (see below).
The CHARMM-GUI webserver (https://www.charmm-gui.org/) was used to assign the side-chain protonation states and embed the models in a bilayer of POPC and cholesterol in 3:1 ratio. The systems were solvated in a periodic box of explicit water and neutralized with 0.15 M of Na+ and Cl− ions. We used the Amber ff14SB GAFF2 and lipid21 force fields, the TIP3P water model, and the Joung-Cheatham ion parameters. Effective point charges of the ligands were obtained by RESP fitting of the electrostatic potentials calculated with the HF/6-31G* basis set using Gaussian 16.
REST2 MD.
After energy minimization, all-atom REST2 MD simulations were carried out using Gromacs2020 patched with PLUMED2.3. The LINCS algorithm was applied to constrain bonds involving hydrogen atoms, allowing for a time step of 2 fs. Each system was gradually heated to 310 K and pre-equilibrated during 10 ns of brute-force MD in the NPT-ensemble. REST2 MD were then performed with a total of 96 replicas in the NVT ensemble. REST2 is a type of Hamiltonian replica exchange simulation scheme (SI Appendix, Fig. S8A). Besides the original simulation, many replicas of the same system were simulated simultaneously. The additional replicas have modified free energy surfaces, in which the energy barriers are easier to cross than in the original simulation system. By frequently swapping the replicas and the original system during the MD, the simulations “travel” on different free energy surfaces and easily visit various conformational zones. Finally, only the samples on the original free energy surface are collected. The additional replicas are artificial to ease barrier crossing, which are discarded after the simulations. REST2, in particular, modifies the free energy surfaces by scaling (reducing) the force constants of the “solute” molecules in the simulation system. In this case, the protein was considered as solute—the force constants of its van der Waals, electrostatic, and dihedral terms were subject to scaling in order to facilitate the conformational changes. The scaling factors were generated using the Patriksson-van der Spoel approach (79) and effective temperatures ranging from 310 K to 1,800 K. Exchange between replicas was attempted every 1,000 simulation steps. This setup resulted in an average exchange probability of ~30%. The high temperatures were used to enhance the sampling. Although they may lead to protein misfolding, such unrealistic conformations were rejected during the exchanges. Finally, he simulation trajectories on the original unmodified free energy surface were reassembled and analyzed.
Several independent REST2 MD runs were performed on different docked poses of each ligand until a stable binding pose was obtained, in which the ligand rmsd was <2 Å during at least 30 ns. Discarding the unstable binding poses, the remaining simulations were prolonged until the HDX profile could be reproduced. Some of the binding poses appeared stable during the MD simulations but the calculated ΔlnPF values were incoherent with the HDX profile (see SI Appendix, Fig. S9 for an example), which were also discarded. These were likely due to insufficient MD sampling in which bulky ligands could be “stuck” at a local minimum of irrelevant binding poses. The comparison with the HDX-MS data thus served both to exclude the false binding poses and to assess the accuracy of the final prediction. In other words, we used the HDX-MS data to guide and validate the REST2 MD in a trial-and-error process. A total of >1.5 ms of REST2 MD simulations (including replicas) were performed. The final production runs contained 100 ns × 96 replicas for the agonist-bound systems and 60 ns × 96 replicas for the other systems that were less dynamic (SI Appendix, Fig. S10A). The last 30 ns were used for HDX PF calculations using HDXer (43). For the inverse agonists, the trajectories of both enantiomers (30 ns each) were included. lnPF of each residue i was calculated as (80), where NC,i the number of nonhydrogen atoms within 6.5 Å of the backbone amide N atom of residue i (excluding atoms in residues i − 2 to i + 2); NH,i is the number of O or N atoms within 2.4 Å of the amide hydrogen atom of residue i; and indicates the ensemble average of the MD simulation frames used. All the atoms of the simulation system, with the exception of water molecules, were taken into account. The scaling factors βC and βH were set to their default values. Note that we did not use the reweighting function of HDXer but its basic implementation of the Best-Vendruscolo method (80), which calculates lnPFi from input MD trajectories directly (81). ΔlnPFi was then calculated as the difference from apo ACKR3, namely ΔlnPFi = lnPFi − lnPFi,apo. Finally, for comparison with the HDX-MS data, ΔlnPFi of residue i was averaged across all the peptides obtained in HDX-MS: , in which mj and nj are the first and last residue numbers of the jth peptide, and ⟨⋯⟩ indicates the average across all the peptides to calculate the final for residue i plotted in Figs. 2 and 3 (Dataset S3). Proline residues and the first residue mj in each peptide were excluded in the calculations. Note that the comparison between the calculated ΔlnPF and the measured ΔRFU was qualitative.
Principal component analysis was performed with bio3d, after an alignment on the most invariant region of the receptor. The most invariant region was identified by iterations of structural superposition, excluding the most variable regions after each round.
Reservoir replica exchange MD (R-REMD).
Additional simulations were performed suing R-REMD to assess the reproducibility of the above REST2 MD sampling, as well as the general applicability of the two methods to other systems. Although REST2 MD has a remarkable capability of conformational exploration, it is often challenging to achieve convergence (discussed in SI Appendix, Supplementary Methods and Fig. S10A). R-REMD is a variant of replica exchange MD (SI Appendix, Fig. S8B) that further accelerates sampling and convergence (46, 47). The recent implementation of GPU-acceleration and thorough assessment of the technique (46) enabled R-REMD of large systems like GPCRs, although it currently only allows for temperature replica exchange. R-REMD simulations were performed using Amber20 and the same force fields as above. A reservoir of MD trajectory was precomputed for each of the four systems to initiate R-REMD (see SI Appendix, Supplementary Methods for the reservoir generation). A clustering analysis was performed on the reservoir to examine convergence (SI Appendix, Fig. S10B), using Amber Cpptraj and the hierarchical agglomerative algorithm. The default average-linkage method was used for clustering on the Cα atoms of the transmembrane helices and ICL2 with a cutoff of 1.5 Å, eliminating clusters that contained <1% of the trajectory. From the representative structures of the top two clusters, two independent R-REMD runs were initiated for each system, coupled to the respective reservoir. Each R-REMD run was carried out in two steps in a multiple R-REMD approach (82), which significantly accelerates sampling and demands fewer parallel computing processes. Step 1 used the reservoir generated above (non-Boltzmann-weighted reservoir) and a temperature range of 370 K to 450 K. Subsequently, step 2 used the trajectory of the 370 K replica from step 1 as a (Boltzmann-weighted) reservoir and a temperature range of 310 K to 370 K. Considering the available computing resources, we chose 48 replicas for each step to achieve ~40% exchange rate between replicas and ~15 ns/d/replica (on 4 NVIDIA® Tesla® V100 GPUs and 144 Intel® Cascade Lake CPU cores). Each step underwent 35 ns × 48 replicas and the first five ns were discarded for equilibration. A total of 26.88 µs of R-REMD simulations were performed for the four systems, from which ΔlnPF values were calculated from the lowest-temperature (310 K) replica.
The two independent R-REMD runs gave consistent results with the HDX-MS and the REST2 MD data (SI Appendix, Fig. S11). Discrepancies in TM7-H8 of the agonist-bound form persisted, due to overestimation of the ionic lock in apo ACKR3, as discussed for the REST2 MD. The R-REMD required fewer trial-and-error interventions, resulting in more conformational clusters, better convergence (SI Appendix, Fig. S10B), and greater reproducibility. However, the initial setup of the R-REMD here relied on the knowledge gained from the prior REST2 MD (see SI Appendix, Supplementary Methods for more details). Therefore, although both methods are readily applicable to other systems, their effectiveness may vary from one case to another and remains to be investigated in future work.
BRET-Based β-Arrestin1 Recruitment Assay.
HEK293T cells were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C with 5% CO2. For β-arrestin recruitment, cells were seeded at a density of 30,000 cells/well and transiently transfected with 4 ng/well of Flag-Snap-ACKR3-NLuc or relevant mutants and 40 ng/well of β-arrestin1-YFP using lipofectamine 2000 (Life technologies). 24 to 48 h post transfection, cells were washed once with PBS and subsequently treated with 40 μL of CXCL12 (RnD Systems) serially diluted in PBS containing 0.1% Bovine Serum Albumin (Sigma). After 15 min of incubation at 37 °C, 5% CO2, 5 µM Colenterazine H was added and BRET was measured on a Tecan infinite F500. BRET ratio was calculated by dividing the YFP emission (535 nm) by Nluc (460 nm). Dose–response curves were generated in GraphPad Prism 9.5.1 (GraphPad Software, Inc.) using the equation “log (agonist) vs. response.” All tested constructs showed comparable EC50s, for similar expression of the receptors, suggesting that CXCL12 has equal potency on the mutant and wild type receptors.
TR-FRET-Based CXCL12 Binding Assay.
HEK293 cells were transiently transfected into 96-well with 2 ng/well of Flag-Snap-ACKR3-NLuc or relevant mutants and 40 ng/well of β-arrestin1-YFP using lipofectamine 2000. 24 to 48 h posttransfection, cells were washed once with tag-lite buffer (Revvity) and labeled with Snap-Lumi4-Tb (100 nm; Revvity) for 1 h at room temperature. Cells were subsequently incubated with CXCL12 serially diluted in tag-lite buffer for 15 min at room temperature followed by incubation with CXCL12-red (1.5 nM) for 15 min at 37 °C, 5% CO2. Data were collected on a Pherstar (BMG Labtech) with excitation at 335 nm, terbium emission at 620 nm (donor) and 665 nm (TR-FRET) over time. The ratio of the signals (665/620) was plotted and analyzed using GraphPad Prism 9.5.1 using the equation “log (inhibitor) vs. response-variable slope.” As all tested constructs showed comparable IC50s, we assume equal affinity of CXCL12 at mutant receptors compared to wildtype.
Analysis of ACKR3 Ligand Binding Site by Site-Directed Mutagenesis.
The binding sites of the agonist VUF15485 and the inverse agonist VUF16840 were also probed by site-directed mutagenesis. N-terminally HA-tagged ACKR3 mutants, D1794.60N, E2135.39Q, D2756.58N, Q3017.39E, and Q3017.39A were generated by PCR-based mutagenesis, sequence-verified, and expressed in HEK293T cells, as described previously (34). The binding of VUF15485 and VUF16840 to wild-type and mutant ACKR3 was measured in [3H]VUF15485 (in-house synthesized) radioligand binding studies (34). Briefly, membranes expressing the wild type or mutant HA-ACKR3 were incubated with [3H]VUF15485 and increasing concentrations VUF16840 or VUF15485. All dilutions were prepared in HBSS supplemented with 0.2% BSA. Membranes and radioligands were optimized to measure specific binding of [3H]VUF15485 to the ACKR3 and mutants, with 1,000 to 5,000 dpm. Membrane protein contents ranged between 4 μg and 80 μg per well and [3H]VUF15485 ranged between 2 nM and 20 nM. All conditions were performed in triplicate on a 96-well plate. Binding reactions were incubated for 2 h and then terminated by washing the solutions over a PEI-coated GF/C filter using a cell harvester (Perkin Elmer) followed by wash steps using ice-cold buffer [50 mM HEPES, 1.2 mM CaCl2, 5 mM MgCl2, 0.5 M NaCl, pH 7.4 at 4 °C]. Filter-bound [3H]VUF15485 was measured by adding 25 µL Microscint-O per well to the dried GF/C-plate and radioactivity was consecutively quantified using the Wallac Microbeta counter (Perkin Elmer). The sigmoidal dose-dependent displacement curves of [3H]VUF15485 binding by unlabeled ligands were analyzed using Graphpad Prism 8, by fitting the data to a one-site binding model to determine the pIC50. Binding affinities (Ki values) for VUF15485 and VUF16840 were determined using the Cheng-Prusoff equation (83).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Acknowledgments
This research was funded by European Union (H2020-MSCA Program, Grant agreement 860229-ONCORNET2.0 and Grant Agreement 641833-ONCORNET). Technical assistance of Drs. Simon Mobach is highly valued. Mass spectrometry experiments were carried out using the facilities of the Pôle Protéome de Montpellier (BioCampus Montpellier). This work was supported by the European Regional Development Fund/Région Occitanie, Montpellier Université d’Excellence, Labex EpiGenMed, and the French Agence Nationale de la Recherche (projects LEUKOCEPTOR ANR-21-CE44-0007-01 and OCHRE no R19168LL). MD simulations were performed using HPC resources from GENCI-TGCC France.
Author contributions
X.C. and C.B. designed research; O.O., C.A., E.D.N., M.A.-S., R.B., B.Z., T.G., C.L., C.d.G., R.L., X.C., and C.B. performed research; O.O., C.A., E.D.N., M.A.-S., R.B., B.Z., T.G., C.L., C.d.G., R.L., T.D., S.G., X.C., and C.B. analyzed data; and O.O., X.C., and C.B. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Xiaojing Cong, Email: xiaojing.cong@igf.cnrs.fr.
Cherine Bechara, Email: cherine.bechara@igf.cnrs.fr.
Data, Materials, and Software Availability
The HDX mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD044103 (84). All other data are included in the article and/or supporting information.
Supporting Information
References
- 1.Griffith J. W., Sokol C. L., Luster A. D., Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Bonecchi R., Graham G. J., Atypical chemokine receptors and their roles in the resolution of the inflammatory response. Front. Immunol. 7, 224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balabanian K., et al. , The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280, 35760–35766 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Meyrath M., et al. , The atypical chemokine receptor ACKR3/CXCR7 is a broad-spectrum scavenger for opioid peptides. Nat. Commun. 11, 3033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smit M. J., et al. , The CXCL12/CXCR4/ACKR3 axis in the tumor microenvironment: Signaling, crosstalk, and therapeutic targeting. Annu. Rev. Pharmacol. Toxicol. 61, 541–563 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Canals M., et al. , Ubiquitination of CXCR7 controls receptor trafficking. PLoS One 7, e34192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajagopal S., et al. , β-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc. Natl. Acad. Sci. U.S.A. 107, 628–632 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen Y.-C., et al. , Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias. Sci. Adv. 8, eabn8063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleist A. B., et al. , Conformational selection guides β-arrestin recruitment at a biased G protein–coupled receptor. Science 377, 222–228 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballesteros J. A., Weinstein H., Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995). [Google Scholar]
- 11.Zhang W., et al. , A point mutation that confers constitutive activity to CXCR4 reveals that T140 is an inverse agonist and that AMD3100 and ALX40-4C are weak partial agonists. J. Biol. Chem. 277, 24515–24521 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Gusach A., et al. , Beyond structure: Emerging approaches to study GPCR dynamics. Curr. Opin. Struct. Biol. 63, 18–25 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Weis W. I., Kobilka B. K., The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Congreve M., De Graaf C., Swain N. A., Tate C. G., Impact of GPCR structures on drug discovery. Cell 181, 81–91 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Van Drie J. H., Tong L., Cryo-EM as a powerful tool for drug discovery. Bioorg. Med. Chem. Lett. 30, 127524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong X., et al. , Molecular insights into the biased signaling mechanism of the μ-opioid receptor. Mol. Cell 81, 4165–4175.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erlandson S. C., et al. , The relaxin receptor RXFP1 signals through a mechanism of autoinhibition. Nat. Chem. Biol. 19, 1013–1021 (2023), 10.1038/s41589-023-01321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heng J., et al. , Function and dynamics of the intrinsically disordered carboxyl terminus of β2 adrenergic receptor. Nat. Commun. 14, 2005 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu Q., et al. , Insights into distinct signaling profiles of the µOR activated by diverse agonists. Nat. Chem Biol 19, 423–430 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang Y., et al. , Molecular recognition of morphine and fentanyl by the human μ-opioid receptor. Cell 185, 4361–4375.e19 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Grison C. M., et al. , Molecular insights into mechanisms of GPCR hijacking by Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 118, e2108856118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josephs T. M., et al. , Structure and dynamics of the CGRP receptor in apo and peptide-bound forms. Science 372, eabf7258 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Lee S.-M., et al. , Calcitonin receptor N-glycosylation enhances peptide hormone affinity by controlling receptor dynamics. J. Mol. Biol. 432, 1996–2014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West G. M., et al. , Ligand-dependent perturbation of the conformational ensemble for the GPCR β2 adrenergic receptor revealed by HDX. Structure 19, 1424–1432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L., et al. , Conformational states of the full-length glucagon receptor. Nat. Commun. 6, 7859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., et al. , Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer. Structure 18, 1332–1341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du Y., et al. , Assembly of a GPCR-G protein complex. Cell 177, 1232–1242.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H. R., et al. , Structural mechanism underlying primary and secondary coupling between GPCRs and the Gi/o family. Nat. Commun. 11, 3160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodge E. A., Benhaim M. A., Lee K. K., Bridging protein structure, dynamics, and function using hydrogen/deuterium-exchange mass spectrometry. Protein Sci. 29, 843–855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James E. I., Murphree T. A., Vorauer C., Engen J. R., Guttman M., Advances in hydrogen/deuterium exchange mass spectrometry and the pursuit of challenging biological systems. Chem. Rev. 122, 7562–7623 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia R., Bradshaw R. T., Calvaresi V., Politis A., Integrating hydrogen deuterium exchange-mass spectrometry with molecular simulations enables quantification of the conformational populations of the sugar transporter XylE. J. Am. Chem. Soc. 145, 7768–7779 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaptal V., et al. , Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter. Sci. Adv. 8, eabg9215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wijtmans M., et al. , Synthesis, modeling and functional activity of substituted styrene-amides as small-molecule CXCR7 agonists. Eur. J. Med. Chem. 51, 184–192 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Zarca A., et al. , Pharmacological characterization and radiolabeling of VUF15485, a high-affinity small-molecule agonist for the atypical chemokine receptor ACKR3. Mol. Pharmacol. 105, 301–312 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Bosma R., et al. , Inactivation of the ACKR3 by a small-molecule inverse agonist. iGPCRnet. http://www.i-gpcrnet.com/file/medtool/webmedtool/cnrstool01/botm0039/pdf00002.pdf. Accessed 25 June 2023.
- 36.Pouzol L., et al. , ACT-1004-1239, a first-in-class CXCR7 antagonist with both immunomodulatory and promyelinating effects for the treatment of inflammatory demyelinating diseases. FASEB J. 35, e21431 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Richard-Bildstein S., et al. , Discovery of the potent, selective, orally available CXCR7 antagonist ACT-1004-1239. J. Med. Chem. 63, 15864–15882 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Wang L., Friesner R. A., Berne B. J., Replica exchange with solute scaling: A more efficient version of replica exchange with solute tempering (REST2). J. Phys. Chem. B 115, 9431–9438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Healey R. D., et al. , Discovery and mechanism of action of small molecule inhibitors of ceramidases. Angew. Chem. Int. Ed. Engl. 61, e202109967 (2021), 10.1002/anie.202109967. [DOI] [PubMed] [Google Scholar]
- 40.Cong X., Fiorucci S., Golebiowski J., Activation dynamics of the neurotensin G protein-coupled receptor 1. J. Chem. Theory Comput. 14, 4467–4473 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Cong X., Golebiowski J., Allosteric Na+ -binding site modulates CXCR4 activation. Phys. Chem. Chem. Phys. 20, 24915–24920 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Cong X., Chéron J.-B., Golebiowski J., Antonczak S., Fiorucci S., Allosteric modulation mechanism of the mGluR5 transmembrane domain. J. Chem. Inf. Model. 59, 2871–2878 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Bradshaw R. T., Marinelli F., Faraldo-Gómez J. D., Forrest L. R., Interpretation of HDX data by maximum-entropy reweighting of simulated structural ensembles. Biophys. J. 118, 1649–1664 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Q., et al. , Common activation mechanism of class A GPCRs. Elife 8, e50279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naumann U., et al. , CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One 5, e9175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kasavajhala K., Lam K., Simmerling C., Exploring protocols to build reservoirs to accelerate temperature replica exchange MD simulations. J. Chem. Theory Comput. 16, 7776–7799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roitberg A. E., Okur A., Simmerling C., Coupling of replica exchange simulations to a Non-Boltzmann structure reservoir. J. Phys. Chem. B 111, 2415–2418 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lau A. M., Claesen J., Hansen K., Politis A., Deuteros 2.0: Peptide-level significance testing of data from hydrogen deuterium exchange mass spectrometry. Bioinformatics 37, 270–272 (2020), 10.1093/bioinformatics/btaa677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hauser A. S., et al. , GPCR activation mechanisms across classes and macro/microscales. Nat. Struct. Mol. Biol. 28, 879–888 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang Y., et al. , Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y., et al. , Molecular basis of β-arrestin coupling to formoterol-bound β1-adrenoceptor. Nature 583, 862–866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schafer C. T., Pauszek R. F., Gustavsson M., Handel T. M., Millar D. P., Distinct activation mechanisms of CXCR4 and ACKR3 revealed by single-molecule analysis of their conformational landscapes. bioRxiv [Preprint] (2023). 10.1101/2023.10.31.564925v1 (Accessed 24 November 2023). [DOI]
- 53.Suno R., et al. , Structural insights into the G protein selectivity revealed by the human EP3-Gi signaling complex. Cell Rep. 40, 111323 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Cao D., et al. , Structure-based discovery of nonhallucinogenic psychedelic analogs. Science 375, 403–411 (2022). [DOI] [PubMed] [Google Scholar]
- 55.McCorvy J. D., et al. , Structural determinants of 5-HT2B receptor activation and biased agonism. Nat. Struct. Mol. Biol. 25, 787–796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shao Z., et al. , Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1. Nat. Chem. Biol. 18, 264–271 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asher W. B., et al. , GPCR-mediated β-arrestin activation deconvoluted with single-molecule precision. Cell 185, 1661–1675.e16 (2022), 10.1016/j.cell.2022.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katritch V., et al. , Allosteric sodium in class A GPCR signaling. Trends Biochem. Sci. 39, 233–244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bous J., et al. , Structure of the vasopressin hormone–V2 receptor–β-arrestin1 ternary complex. Sci. Adv. 8, eabo7761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin W., et al. , A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 29, 971–983 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Staus D. P., et al. , Structure of the M2 muscarinic receptor–β-arrestin complex in a lipid nanodisc. Nature 579, 297–302 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W., et al. , Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 579, 303–308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duval V., Alayrac P., Silvestre J.-S., Levoye A., Emerging roles of the atypical chemokine receptor 3 (ACKR3) in cardiovascular diseases. Front. Endocrinol. 13, 906586 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bayrak A., et al. , Discovery and development of first-in-class ACKR3/CXCR7 superagonists for platelet degranulation modulation. J. Med. Chem. 65, 13365–13384 (2022), 10.1021/acs.jmedchem.2c01198. [DOI] [PubMed] [Google Scholar]
- 65.Miao Z., et al. , CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc. Natl. Acad. Sci. U.S.A. 104, 15735–15740 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cvicek V., Goddard W. A., Abrol R., Structure-based sequence alignment of the transmembrane domains of all human GPCRs: Phylogenetic, structural and functional implications. PLoS Comput. Biol. 12, e1004805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levoye A., Balabanian K., Baleux F., Bachelerie F., Lagane B., CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood 113, 6085–6093 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Fumagalli A., et al. , The atypical chemokine receptor 3 interacts with Connexin 43 inhibiting astrocytic gap junctional intercellular communication. Nat. Commun. 11, 4855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kahsai A. W., et al. , Signal transduction at GPCRs: Allosteric activation of the ERK MAPK by β-arrestin. Proc. Natl. Acad. Sci. U.S.A. 120, e2303794120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomsen A. R. B., et al. , GPCR-G protein-β-arrestin super-complex mediates sustained G protein signaling. Cell 166, 907–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Q., et al. , ACKR3–arrestin2/3 complexes reveal molecular consequences of GRK-dependent barcoding. bioRxiv [Preprint] (2023). 10.1101/2023.07.18.549504 (Accessed 19 July 2023). [DOI]
- 72.Schönegge A.-M., et al. , Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity. Nat. Commun. 8, 2169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wingler L. M., et al. , Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR. Science 367, 888–892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu Z., et al. , Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism. Nat. Chem. Biol. 18, 281–288 (2022). [DOI] [PubMed] [Google Scholar]
- 75.Qu Q., et al. , Insights into distinct signaling profiles of the µOR activated by diverse agonists. Nat. Chem. Biol. 19, 423–430 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suomivuori C.-M., et al. , Molecular mechanism of biased signaling in a prototypical G protein–coupled receptor. Science 367, 881–887 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benredjem B., Girard M., Rhainds D., St.-Onge G., Heveker N., Mutational analysis of atypical chemokine receptor 3 (ACKR3/CXCR7) interaction with its chemokine ligands CXCL11 and CXCL12. J. Biol. Chem. 292, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patriksson A., Van Der Spoel D., A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys. 10, 2073 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Best R. B., Vendruscolo M., Structural interpretation of hydrogen exchange protection factors in proteins: Characterization of the native state fluctuations of CI2. Structure 14, 97–106 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Lee P. S., et al. , Interpreting hydrogen-deuterium exchange experiments with molecular simulations: Tutorials and applications of the HDXer ensemble reweighting software [Article v1.0]. Living J. Comput. Mol. Sci. 3, 1521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ruscio J. Z., Fawzi N. L., Head-Gordon T., How hot? Systematic convergence of the replica exchange method using multiple reservoirs. J. Comput. Chem. 31, 620–627 (2009), 10.1002/jcc.21355. [DOI] [PubMed] [Google Scholar]
- 83.Cheng Y., Prusoff W. H., Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973). [DOI] [PubMed] [Google Scholar]
- 84.PRIDE Archive, Conformational dynamics underlying Atypical Chemokine Receptor 3 activation. PRoteomics IDEntifications Database (PRIDE). https://www.ebi.ac.uk/pride/archive/projects/PXD044103/private. Deposited 27 July 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Data Availability Statement
The HDX mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD044103 (84). All other data are included in the article and/or supporting information.