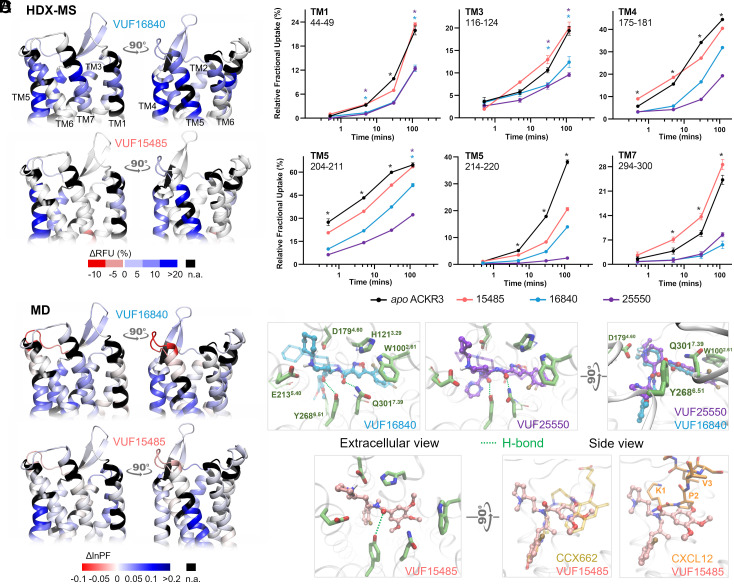
Fig. 2.
Ligand binding at the level of ACKR3 orthosteric pocket. (A) Schematic representation of the % differential relative fractional uptake data (apo ACKR3—bound ACKR3) mapped onto the upper part of the AF model of ACKR3 (for clarity, we did not include the N terminus). Relative fractional uptake is calculated by dividing the experimental uptake (Da) of a peptide by its maximum possible uptake. This depicts reproducible and statistically significant ΔHDX in response to inverse agonists small ligands represented by VUF16840 (VUF25550 giving a similar profile), or agonist small ligand VUF15485. Black regions represent regions with no sequence coverage. Ligand-induced reduction in deuterium uptake is represented in blue while ligand-induced increase in deuterium uptake is in red, according to the scale. (B) Associated deuterium uptake plots showing the relative uptake for peptides from apo or ligand-bound ACKR3 across several deuteration time points and are representative of the extracellular region as indicated at the Top of the plot. Statistically significant changes were determined using Deuteros 2.0 software (48) (P ≤ 0.01): statistically significant time points for the different ligands are represented by a colored star corresponding to the ligand in question (pink, blue, and purple for VUF15485, VUF16840, and VUF25550 respectively). Black stars depict time points that are statistically significant for all three ligands. Uptake plots are the average and SD of three technical replicates from the same biological preparation of ACKR3. (C) Calculated differential ln HDX protection factor changes (ΔlnPF) mapped on the AF model. Per-residue lnPF was first calculated for each MD trajectory of ACKR3 in apo and ligand-bound forms. The difference between the apo and bound forms gave the per-residue ΔlnPF for each ligand. For comparison with the HDX data, per-peptide ΔlnPF was calculated by averaging the per-residue ΔlnPF over the peptides obtained in the HDX-MS experiments for each ligand (see Materials and Methods and Dataset S3 for details). (D) Predicted ligand binding mode. The inverse agonists could bind in both enantiomers of the piperidine group (solid and transparent depictions). They showed nearly identical binding poses except that the cyclopropyl-pyrimidine of VUF25550 was more mobile. The agonist VUF15485 formed ionic interactions with D1794.60 via its 1-methylpyrrolidine. The rest of VUF15485 largely overlaps with CCX662 as shown in the superimposition to PDB 7SK8. It also overlaps with the N terminus of CXCL12.