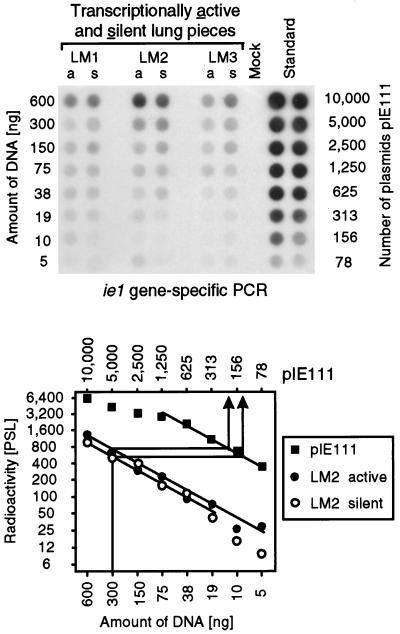
FIG. 2.
Quantitation of the latent viral DNA in transcriptionally active and silent tissue pieces. In parallel with the isolation of the poly(A)+ RNA used for the transcriptional analyses shown in Fig. 1, DNA was isolated from lung tissue pieces #1 through #9 derived from mice LM1 to LM3. For each mouse, DNA pools corresponding to transcriptionally active pieces (pool a) and transcriptionally silent pieces (pool s) were formed, according to the pattern of IE1-specific transcription shown in Fig. 1, lower left. The DNA pools were titrated as indicated and were subjected to an ie1 exon 4-specific PCR. A negative control was provided by DNA isolated from uninfected lungs (Mock). As a standard for the quantitation, this mock DNA was supplemented with plasmid pIE111, titrated in duplicate, and subjected to PCR accordingly. (Top) Autoradiograph of the dot blot obtained after hybridization with a γ-32P-end-labeled internal oligonucleotide probe. (Bottom) Computed phosphorimaging data for the same blot. For the sake of clarity, the computations are depicted as graphs only for mouse LM2. Log-log plots of radioactivity (mean of duplicates in the case of the standard) measured as phosphostimulated luminescence (PSL) units (ordinate) versus the amount of sample DNA (abscissa) are shown. The upper line relates the amount of DNA to the number of plasmids in the pIE111 standard. Calculations were made from the linear portions of the graphs, as shown as an example for 300 ng of DNA containing 200 copies and 120 copies of viral DNA in the active and silent pools, respectively, of mouse LM2. The results, given as copies of the viral genome per 106 lung cells (6 μg of DNA) were as follows: for LM1, a = 1,320 and s = 1,480; for LM2, a = 4,000 and s = 2,400; and for LM3, a = 1,260 and s = 1,460.