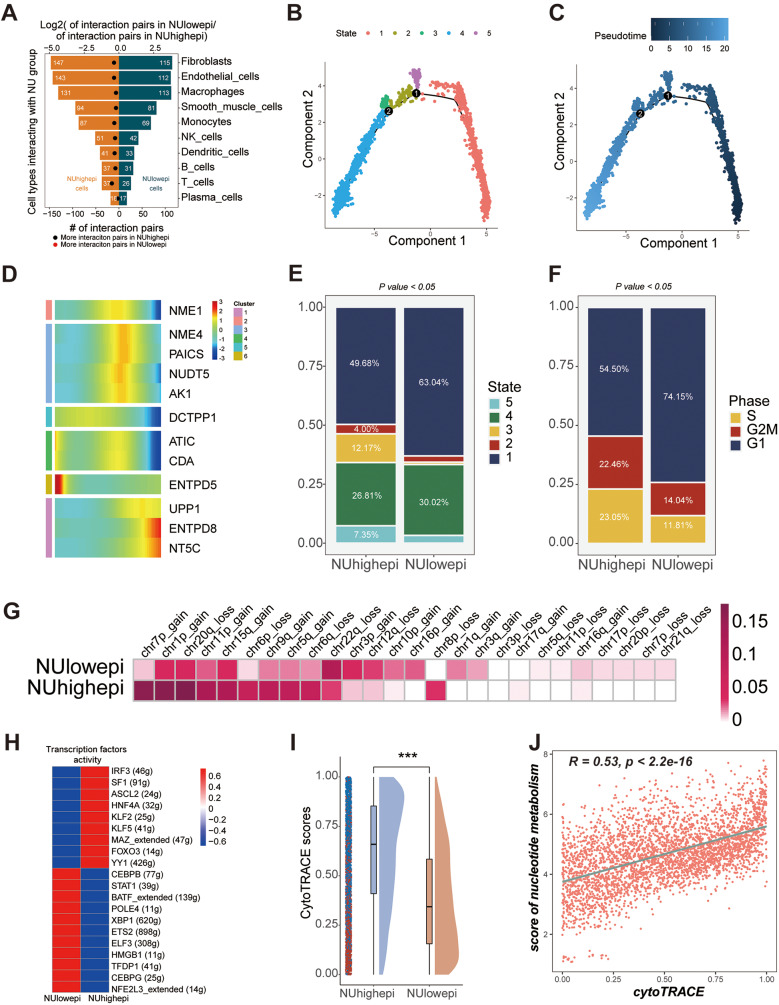
Fig. 3.
Function analysis of NUhighepi/NUlowepi in scRNA-seq data: (A) Barplot showed the number of other cell types communicating with NU group. (B-C) Pseudotime analysis of epithelial cells. (D) Heatmap showed pseudotime of Nucleotide metabolism related genes (NRGs). (E-F) Barplot showed pseudotime states (E) or cell-cycle states (F) of NU group. (G) Heatmap showed inferCNV profiles of NUhighepi and NUlowepi. (H) Heatmap showing transcriptional activity of NU group. Intensity of color indicates average transcription factors activity. (I) Raincloud plot of CytoTRACE scores by NU group in scRNA-seq data. The center of the box plot are median values, the bounds of the box are 25% and 75% quantiles. (J) Scatter plot of the correlation between CytoTRACE score and Nucleotide metabolism score. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns P > 0.05