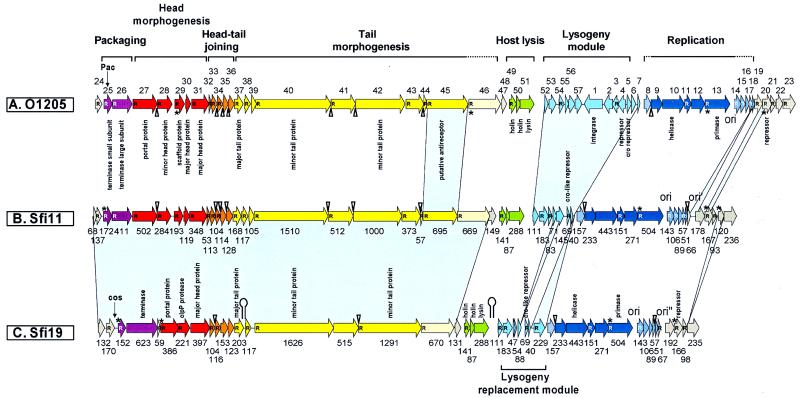
FIG. 1.
Prediction of ORFs in the complete genomes of the temperate pac-site S. thermophilus phage O1205 (top), the virulent pac-site S. thermophilus phage Sfi11 (center), and the virulent cos-site S. thermophilus phage Sfi19 (bottom). To provide a better reference to previous publications, the ORFs from O1205 are numbered as in Stanley et al. (40), while the Sfi11 and Sfi19 ORFs are marked with their lengths in codon numbers. Probable gene functions identified by bioinformatic analysis or biological experiments are noted. The phage genomes were divided into functional units according to previous bioinformatic and comparative evolutionary analysis (32). Genes belonging to the same unit have the same color. Grey shading indicates lack of information about the function of the ORF. ORFs preceded by a potential ribosome binding site (23) are marked with an R inside the arrow. ORFs starting with an unconventional initiation codon are indicated with an asterisk, and possible rho-independent terminators are indicated with a hairpin. The overlap of start and stop codons is indicated with a triangle. Areas of blue shading connect regions of major sequence difference between the compared phage genomes. The genome maps were oriented to maximize alignments; the cos sites and pac sites are thus not at the very ends of the maps.