FIGURE 2. SIRT6 reduces SPTBN1 acetylation.
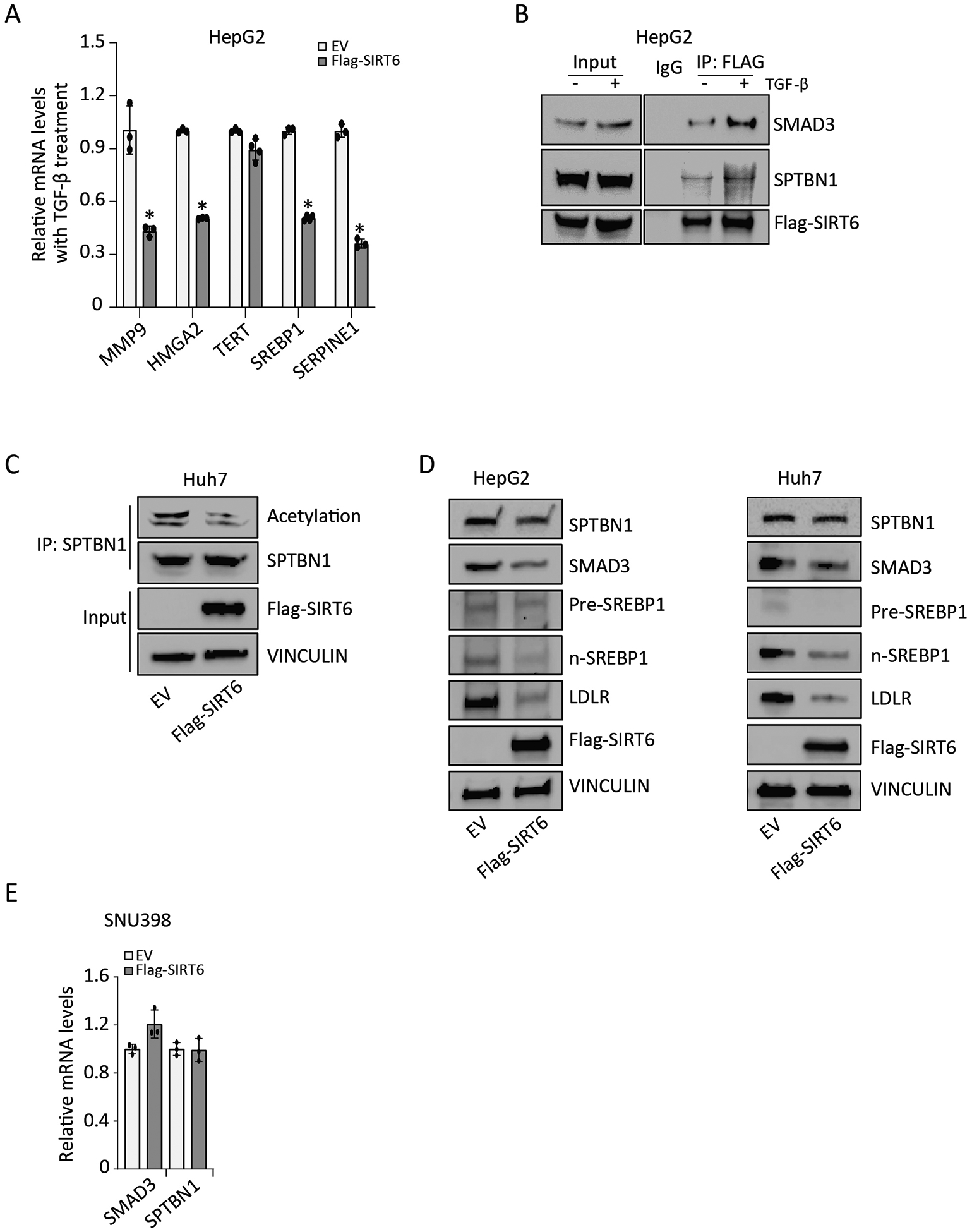
(A) Relative mRNA levels of the indicated TGF-β target genes in HepG2 cells transfected with empty vector (EV) or Flag-SIRT6 and exposed to 200 pM TGF-β for 24 h. Data are presented as mean ± SEM relative to the amount in EV-transfected cells exposed to TGF-β treatment (n = 3 – 4). *, p < 0.05; compared with EV group with TGF-β treatment. (B) Co-immunoprecipitation of SMAD3, SPTBN1, and Flag-SIRT6 was performed in HepG2 cells with or without exposure to 200 pM TGF-β for 3 h, using an irrelevant IgG antibody or anti-FLAG antibody. IP, immunoprecipitation. Representative data from 1 of 3 independent experiments are shown. (C) The effect of SIRT6 overexpression on SPTBN1 acetylation in Huh7 cells was determined by immunoprecipitating SPTBN1 and Western blotting for the presence of acetylation with SPTBN1. Representative data from 1 of 3 independent experiments are shown. (D) The effect of SIRT6 overexpression on SPTBN1, SMAD3, Pre-SREBP1, n-SREBP1, and LDLR abundance in HepG2 cells and Huh7 cells was determined by Western blotting. VINCULIN served as the loading control. EV, empty vector. Representative data from 1 of 3 independent experiments are shown. (E) Relative mRNA levels of SMAD3 and SPTBN1 were determined in SNU398 cells transfected with empty vector (EV) or Flag-SIRT6. Data are presented as mean ± SEM relative to the amount in EV-transfected cells (n = 3). No significant differences were detected with t-test, compared to EV group.
