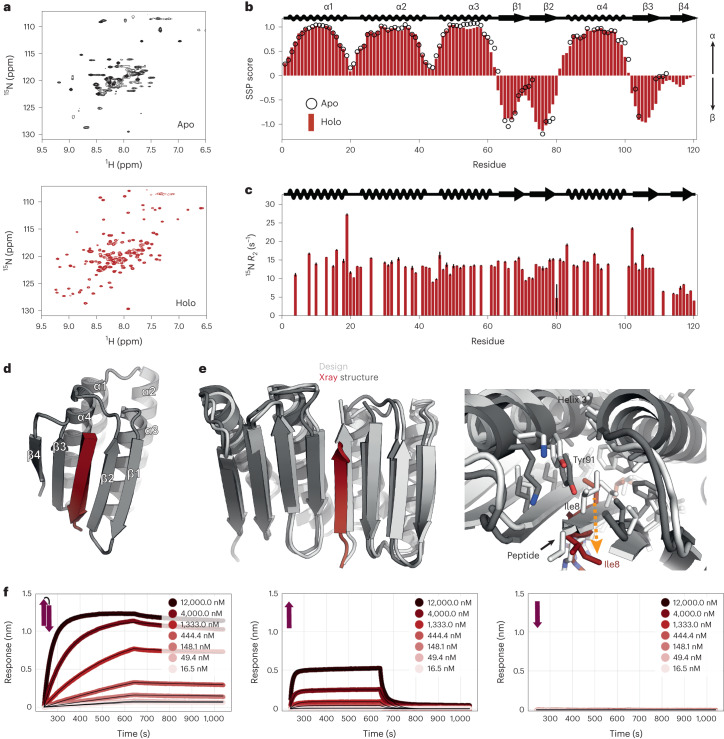
Fig. 4. Structural characterization.
a, NMR spectra of 15N-labeled C34 in the absence (top) and presence (bottom) of tenfold excess target peptide, 25 °C. b, Secondary structure propensity as a function of residue, based on backbone 1H, 13C and 15N chemical shifts recorded at 50 °C using the SSP program34. SSP scores for the apo form are shown with open circles, while those for the peptide-bound state are indicated with bars. The putative secondary structure of the designed protein is indicated above the plot. Positive values of SSP indicate α-helical structure, while negative values denote β-strands. c, 15N transverse relaxation rates as a function of residue. Low values, such as those in putative β4, indicate rapid timescale dynamics and are consistent with poorly formed structure. d, Designed model of C34. e, Left, overlay of the design model of a surface-redesigned version of C104 (gray) and the crystal structure (colors). Right; detailed interface view of the design (gray) and crystal structure (colors) with Ile8 shift indicated by the orange dashed arrow. f, Binding of CH15.1 to its hairpin peptide (left) or to the individual N-terminal strand (middle) or C-terminal strand (right) of the hairpin in BLI.