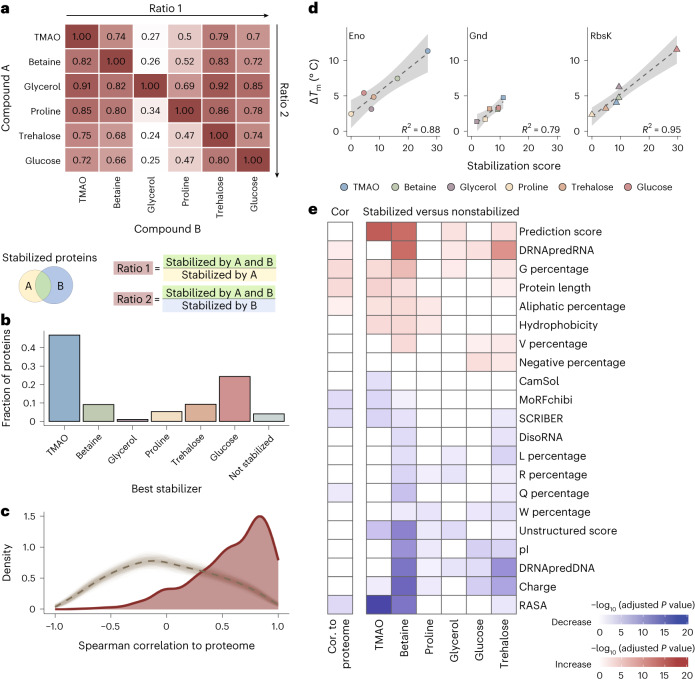
Fig. 3. Biophysical features of osmolyte-stabilized proteins.
a, The heat map shows pairwise overlap of osmolyte-stabilized proteins. Ratio 1 and ratio 2 are calculated as indicated. b, Fraction of proteins stabilized most strongly by each osmolyte out of all proteins detected in all six datasets. c, Density estimate distributions of the Spearman correlation coefficient between the stability score per protein and the mean stability score across the lysate (red line) or a randomized mean stability (dashed gray line) obtained from 1,000 repeated calculations (light gray lines; appears as a gray band). d, Linear regression between DSF-calculated ∆Tm and protein stabilization score for the indicated purified proteins. The shaded area represents the confidence interval of the linear fit (dashed line). Error bars (smaller than point size) show the standard deviation (n = 4 replicates). e, Heat map showing the most significantly different features (adjusted P value of <0.0001 in at least one comparison) between proteins with high correlation and low correlation of the stabilization score with that of the global proteome (first column, Cor) or between proteins significantly stabilized and those not stabilized by the indicated individual osmolytes. The color indicates whether the feature is higher (red) or lower (blue) in proteins with high correlation (first column) and in stabilized proteins (other columns) than in the rest. Feature significance was determined by two-sided t-test followed by correction by multiple hypothesis testing (Benjamini–Hochberg). Other tested features are shown in Supplementary Fig. 3. The term ‘X percentage’ refers to the fraction of the indicated amino acid. See Methods for the calculation or prediction of features.