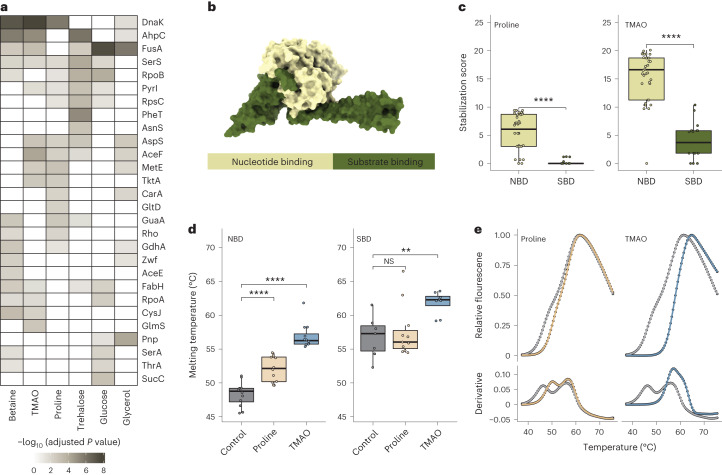
Fig. 4. Stabilization of multidomain proteins.
a, Heat map showing multidomain proteins with domains differentially stabilized by the indicated osmolytes (adjusted P value of <0.05; as determined by two-sided t-test and Benjamini–Hochberg correction). b, Structure of the chaperone DnaK (PDB ID: 4JNE) with the nucleotide binding domain (light green) and substrate binding domain (dark green). c, Distribution of the indicated osmolyte stabilization scores for amino acids mapping to the nucleotide-binding domain (NBD) and substrate-binding domain (SBD) of DnaK. Horizontal lines define the median, and boxes define the 25th and 75th percentiles; whiskers represent the maximum and minimum values. Significance was determined using two-sided Wilcoxon tests; ****P < 0.0001. Each dot represents an amino acid level stabilization score calculated based on two LiP–MS replicates per temperature. d, Distribution of melting temperatures derived from peptides mapping to the indicated domains of DnaK in the presence and absence of the indicated osmolytes. Each point represents one peptide curve measured in duplicate (n ≥ 7 peptides per condition). Box plots are as in c; **P < 0.01; ****P < 0.0001; NS, not significant. e, Protein melting curves measured with DSF and the first derivative of the melting curve for purified DnaK are plotted for control (gray), proline (yellow) and TMAO (blue).