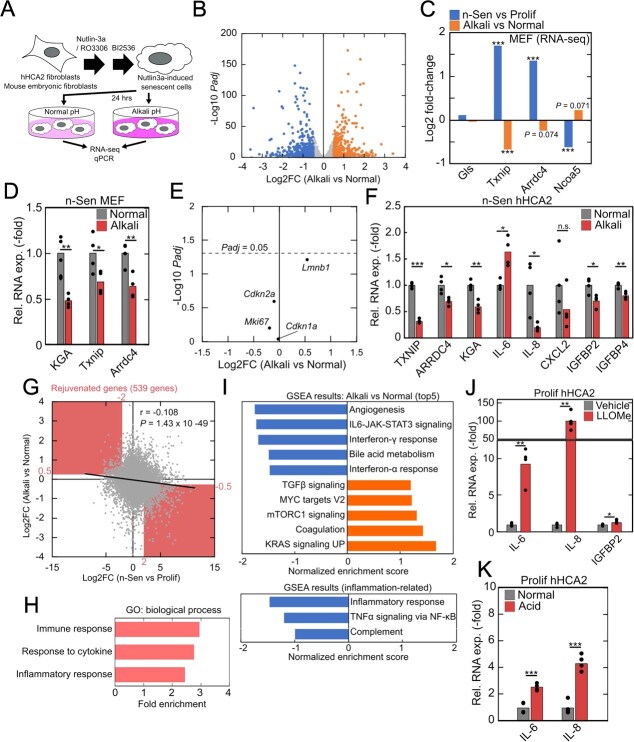
Fig. 2.
Intracellular acidification-dependent gene expression in senescent cells. (A) Schematic illustration of senescence induction and intracellular acid neutralization. (B) Differential gene expression analysis of alkali-treated senescent MEFs. Genes with |Log2FC| > 0.5 and Padj < 0.01 were highlighted. (C-E) Alkali-induced expression changes of pH-responsive genes Gls, Txnip, Arrdc4, and Ncoa5 in n-Sen MEFs (RNA-seq: C, qPCR: D) and senescence-related genes Cdkn2a, Cdkn1a, Mki67, and Lmnb1 (E). (F) qPCR analysis of alkali-induced expression changes of pH-responsive genes in n-Sen hHCA2. (G) Senescence-induced and alkali-induced gene expression changes in MEFs. The significant negative correlation indicated the intracellular acidification dependency of senescence transcriptome. Highlighted area indicates ‘rejuvenated genes’. (H) GO analysis of ‘rejuvenated genes’. (I) GSEA results of differentially expressed genes in response to alkali. Positive values of normalized enrichment score indicate upregulation in alkali-treated senescent MEFs. (J, K) qPCR analysis of gene expression changes in LLOMe-treated (H) and acid-stimulated (I) hHCA2. For qPCR analysis, Student’s t test was used, *P < 0.05; **P < 0.01; ***P < 0.001. For RNA-seq analysis, Benjamini–Hochberg method was used, *Padj < 0.05; **Padj < 0.01; ***Padj < 0.001.