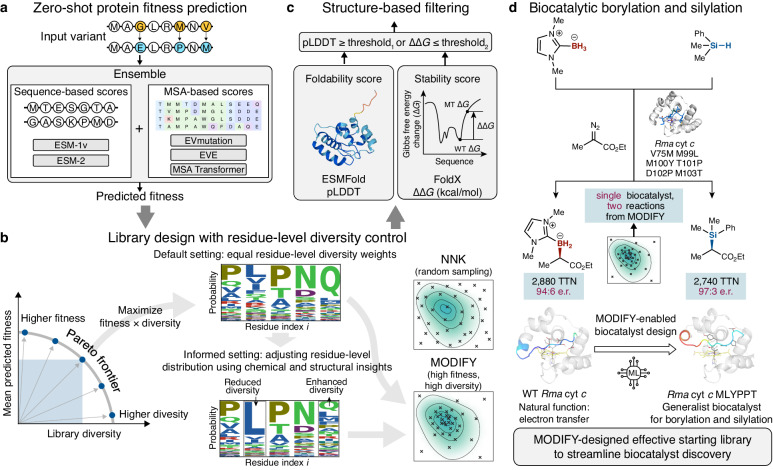
Fig. 1. MODIFY: an ML-guided framework for the design of enzyme engineering starting libraries with both high fitness and high diversity.
a MODIFY leverages pre-trained protein language models and multiple sequence alignment (MSA)-based sequence density models to build an ensemble ML model for zero-shot fitness predictions, effectively eliminating evolutionarily unfavorable variants. b MODIFY co-optimizes the library’s diversity and predicted fitness, pinpointing the Pareto optimal balance between the two. MODIFY offers diversity control at a residue resolution, enabling researchers to either explore a diverse range of amino acids or focus on a subset of compatible amino acids based on biophysical and biochemical insights. c MODIFY further performs a quality control step to filter out problematic variants in the library based on protein foldability (ESMFold pLDDT) and stability (FoldX ΔΔG). d MODIFY-enabled discovery of effective generalist biocatalysts for enantioselective new-to-nature borylation and silylation.