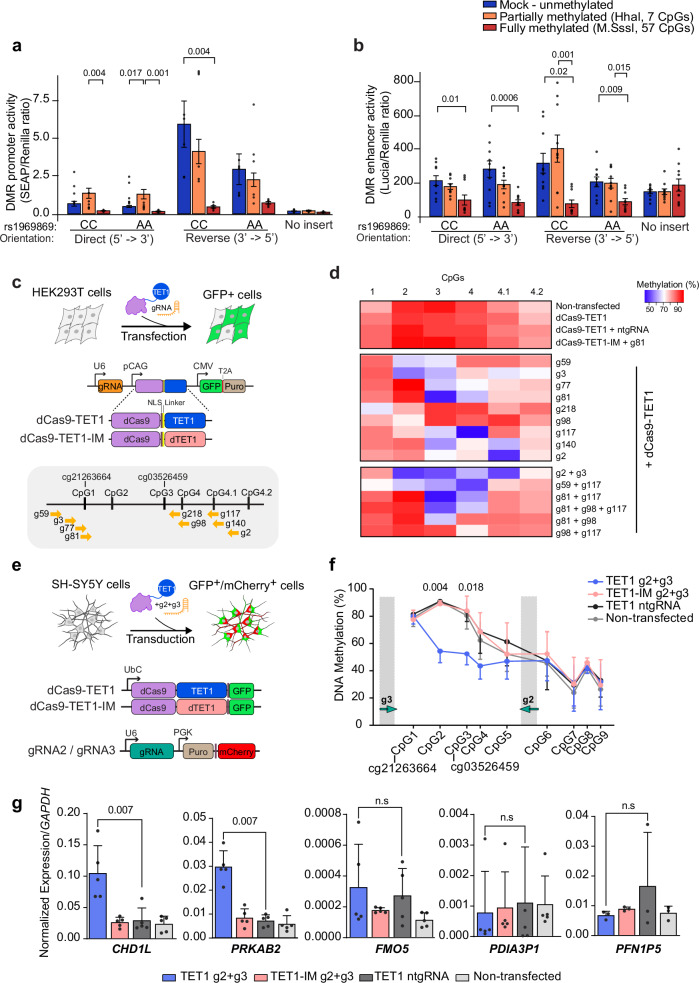
Fig. 4. Functional impact of methylation at the 1q21.1 DMR on gene expression.
a, b Promoter (a) and enhancer (b) activity of the DMR, using CpG-free promoter-free (SEAP) and promoter-containing (Lucia) reporter gene vectors, respectively. Constructs in direct or inverted orientation of DNA segments derived from individuals varying according to the genotype at rs1969869 were partially or fully methylated using HhaI and M.SssI enzymatic treatment, respectively. Results show relative activity ±SEM of SEAP (two experimental replicates, n = 6) or Lucia (three experimental replicates, n = 10) normalized against Renilla (2-way ANOVA with Bonferroni correction for multiple comparisons). c Schematic representation of the experimental design for gRNAs screen in HEK293T cells including the features of the constructs and gRNA locations. d Heatmap of the DNA methylation levels in successfully transfected (GFP positive) HEK293T cells three days following co-transfection of dCas9-TET1 with single or combined gRNAs in comparison to control conditions, deactivated TET1 (TET1-IM), non-targeting gRNA (ntgRNA) and non-transfected cells. e Schematic representation of the experimental design for functional investigation in SH-SY5Y cells. f DNA methylation levels in SH-SY5Y cells following delivery of dCas9-TET1-, gRNA2- and gRNA3-containing lentiviruses. Methylation percentages represent the mean ± SD of three experiments (2-way ANOVA followed by Turkey’s multiple comparisons test). g Experiment showing expression of CHD1L, PRKAB2, PDIA3P1, and FMO5 genes relative to GAPDH transcript levels, quantified using RT-qPCR. The expression levels represent the average of at least three experiments (mean ± SD, two-tailed Student’s t-test). n.s, non-significant. Source data are provided as a Source Data file.