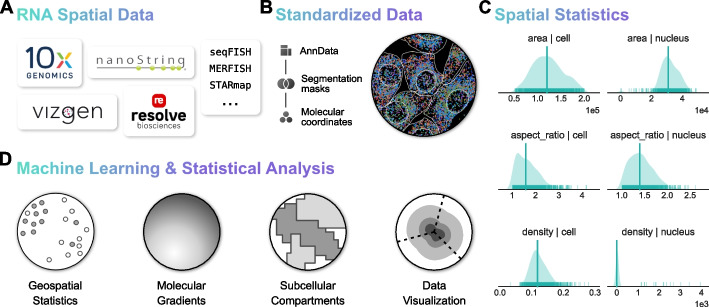
Fig. 1.
Workflow and functionality of the Bento toolkit. A Single-molecule resolved spatial transcriptomics data from commercial or custom platforms are ingested into Bento where it is converted to the AnnData format (B), where it can be manipulated with Bento as well as a wide ecosystem of single-cell omics tools. C Geometric statistics are illustrated for the seqFISH + dataset, including metrics describing cell and nuclear geometries and cell density to assess overall data quality. D Bento has a standard interface to perform a wide variety of subcellular analyses