Extended Data Fig. 1 |. Integration of single cell and nuclei RNA-seq heart datasets during acute responses to MI.
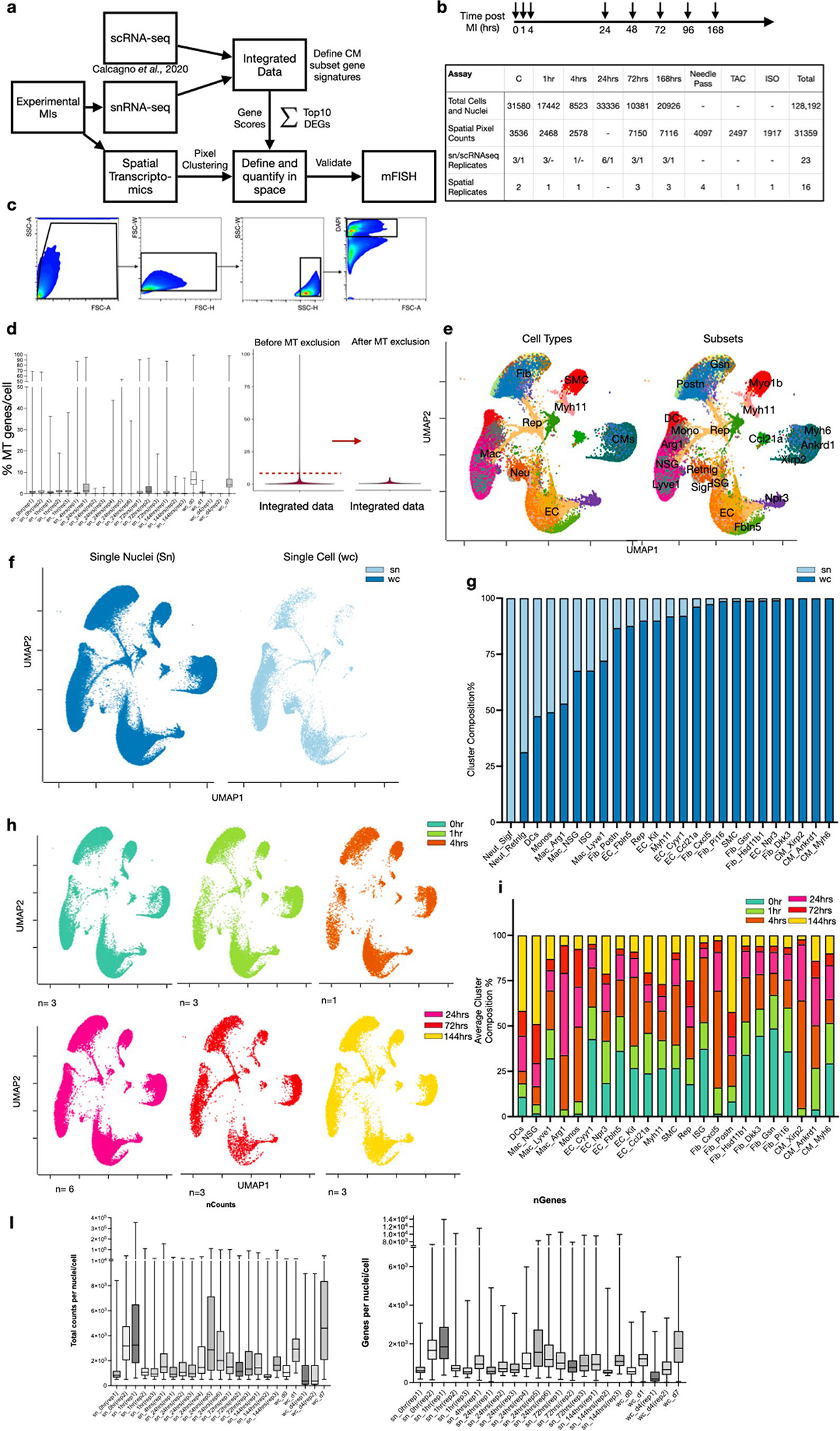
(a) Overall experimental design and integration of sc/snRNA-seq data with whole transcriptome spatial data. Hearts were harvested at several time points following experimental MI and collected for snRNA-seq. The resulting data matrices were integrated with available scRNA-seq data. (b) Experimental timepoints post-MI that were examined in our study with a summary table of total numbers of cells, nuclei and spatial pixels analyzed to support the robustness of our claims across biological replicates. (c) Gating strategy to isolate nuclei using DAPI and FACS. (d) Mitochondrial QC metrics of samples and replicates for both single nuclei, single cell and integrated sn/sc data. (e) UMAP plots annotated by major cell types (left) and subsets (right) after removing nuclei and cells that have more than 5% mitochondrial counts. (f) UMAP plots displaying composition of single nuclei (left) and single cell (right) derived samples. (g) Subcluster composition as derived from UMAPs shown in (f). (h) UMAP plots split by timepoint and across biological replicates. (i) Average subcluster composition displayed in (h). (l) QC metric of samples and replicates for both single nuclei and single cell represented in counts per sample (nCounts) and features per sample (nGenes).
