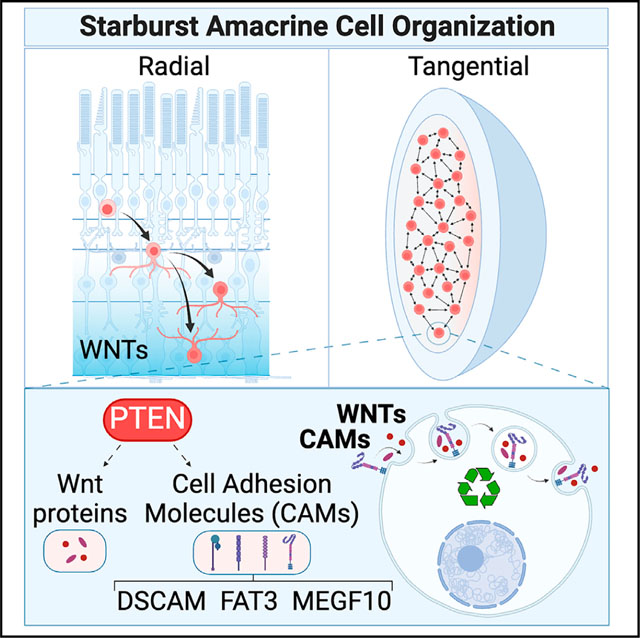
SUMMARY
The retina is exquisitely patterned, with neuronal somata positioned at regular intervals to completely sample the visual field. Here, we show that phosphatase and tensin homolog (Pten) controls starburst amacrine cell spacing by modulating vesicular trafficking of cell adhesion molecules and Wnt proteins. Single-cell transcriptomics and double-mutant analyses revealed that Pten and Down syndrome cell adhesion molecule Dscam) are co-expressed and function additively to pattern starburst amacrine cell mosaics. Mechanistically, Pten loss accelerates the endocytic trafficking of DSCAM, FAT3, and MEGF10 off the cell membrane and into endocytic vesicles in amacrine cells. Accordingly, the vesicular proteome, a molecular signature of the cell of origin, is enriched in exocytosis, vesicle-mediated transport, and receptor internalization proteins in Pten conditional knockout (PtencKO) retinas. Wnt signaling molecules are also enriched in PtencKO retinal vesicles, and the genetic or pharmacological disruption of Wnt signaling phenocopies amacrine cell patterning defects. Pten thus controls vesicular trafficking of cell adhesion and signaling molecules to establish retinal amacrine cell mosaics.
In brief
Patterns in nature range from stereotyped distributions of colored patches on butterfly wings to precise neuronal spacing in the nervous system. Waddington proposed that built-in constraints canalize developmental patterns. Touahri et al. identify Pten-regulated endocytic trafficking of cell adhesion/signaling molecules as a novel constraint measure controlling retinal amacrine cell patterning.
Graphical Abstract
INTRODUCTION
Retinal neurons are organized in two orthogonal dimensions.1 In the radial axis, neurons disperse in an outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL). In the tangential dimension, individual types of retinal neurons further organize into regularly spaced arrays known as mosaics.2 This organization allows for the vertical transmission of individual image points and serial light processing and for the parallel processing of visual information to create a composite map of the visual field.3
Amacrine cells are born at the apical retinal surface and migrate radially to settle in the inner INL or become displaced in the GCL. Radial positioning is regulated by Sox2, which controls the ratio of starburst amacrine cells (SACs), typified by choline acetyltransferase (ChAT) expression, in the GCL versus INL.4 In turn, the FAT atypical cadherin 3 (FAT3) controls bipolar-to-unipolar transitions of radially migrating GABAergic amacrine cells.5 Mosaic establishment and refinements in the tangential plane occur soon after radial layers form, beginning in the first few post-natal days,6–9 with further refinements in the first few weeks.10 Two well-studied mosaics are formed by SACs and dopaminergic amacrine cells characterized by tyrosine hydroxylase (TH) expression, which, respectively, regulate directional sensitivity11 and light adaptation, including transitions between scotopic and photopic vision.12 SACs form archetypical mosaics in both radial layers, establishing exclusion zones that keep them apart from one another.8,13 Such exclusion zones, in the presence of sufficient densities of homotypic cells, ensure regular patterning within the mosaic.14
Amacrine cells use several mechanisms to establish mosaics, including selective cell death (if too close to “like” cells), tangential dispersion (moving away from “like” cells), and selective cell fate specification (altering postmitotic phenotypes if too close to “like” cells).2 Remarkably, each amacrine cell subtype establishes its mosaic pattern independently.13 SACs disperse tangentially,6,15,16 while TH+ amacrine cells use lateral inhibition to block the same fate in amacrine cells spaced too close together during early fate determination,17,18 followed by selective cell death later on, after cells arrive in their strata.19 When horizontal cells are depleted at the time of mosaic formation, these cells space themselves farther apart with larger exclusion zones, indicating that adaptive mechanisms allow complete tiling of the retinal field.20
A computational model of diffusible fate-determining factors that negatively feed back to create exclusion zones can accurately simulate mosaic formation.18,21 Yet, several cell adhesion molecules (CAMs), which act at the cell surface, are essential for mosaic patterning. Indeed, CAMs have a general role in coordinating neuronal positioning and dendritic arborization patterns within neural circuits.22 The type of CAM is important, as is its precise localization, with CAM content tightly regulated by endocytic pathways that traffic CAMs to the cell membrane and remove membrane CAMs to be degraded or transferred to extracellular vesicles (EVs) for secretion.23 For example, in Drosophila, neuroglia is an L1-type CAM that is redistributed from the plasma membrane to endosomes during dendritic pruning.24 Additionally, Down syndrome cell adhesion molecule (DSCAM), which has over 38,000 isoforms in the fly, covers neuronal dendrites, but this coating is reduced during dendritic tiling.25
Murine DSCAM maintains TH+ amacrine cell mosaics by acting as a “non-stick coating,” blocking adhesive signals.26–28 Thus, in Dscam mutants, instead of TH+ amacrine cell dendrites forming continuous nets that uniformly cover the retinal surface, dendrites hyperfasciculate, which secondarily degrades the somal mosaic. Multiple EGF-like domains 10 (MEGF10), a transmembrane CAM, mediates dendrite-soma exclusion to establish SAC somal mosaics.10,29 Furthermore, differential expression of γ-protocadherin isoforms confers SAC self-recognition so that dendrites on the same amacrine cell do not adhere ectopically.30
Phosphatase and tensin homolog (Pten) also controls the radial and tangential positioning of amacrine cells and prevents the fasciculation of TH+ neurites.31 How Pten, a lipid and protein phosphatase that is an eraser and reader of the membrane phospholipid PIP3 (phosphatidyl inositol-3,4,5-triphosphate),32 mediates cell recognition between cells spaced a large distance apart and coordinates non-overlapping dendrite extensions, is unclear. However, PTEN co-localizes with RAB5 in early endosomes33 and dephosphorylates the second messenger PIP3, which directs vesicular formation and transport between subcellular organelles, implicating endocytic processing as a potential intersection point between PTEN and cell-surface molecules.32 Here, we reveal that Pten lies at the top of a cascade of events that controls the endocytic processing of CAMs and Wnt signaling molecules to coordinate SAC mosaic formation.
RESULTS
Pten and Dscam are co-expressed in cholinergic amacrine cells
Pten31 and Dscam34 null mutants partially phenocopy one another, with both displaying aberrant clumping and neurite fasciculation of TH+ amacrine cells. To understand how these molecules interact, we compared their expression profiles at post-natal day 7 (P7), after amacrine cell differentiation is complete and mosaic refinement is in progress.10 In P7 retinas, Dscam mRNA puncta were detected in the INL, where amacrine cell bodies reside, along with bipolar, horizontal, and Müller glial cell somata, and in the GCL, populated by displaced amacrine cells and ganglion cells (Figure 1A). Conversely, Pten transcripts were more widely distributed throughout the INL and GCL (Figure 1A). Higher magnification images confirmed Pten and Dscam transcripts were co-localized in a subset of INL cells (Figure 1A), as were PTEN and DSCAM proteins (Figure 1B).
Figure 1. Pten and Dscam are co-expressed in cholinergic amacrine cells.
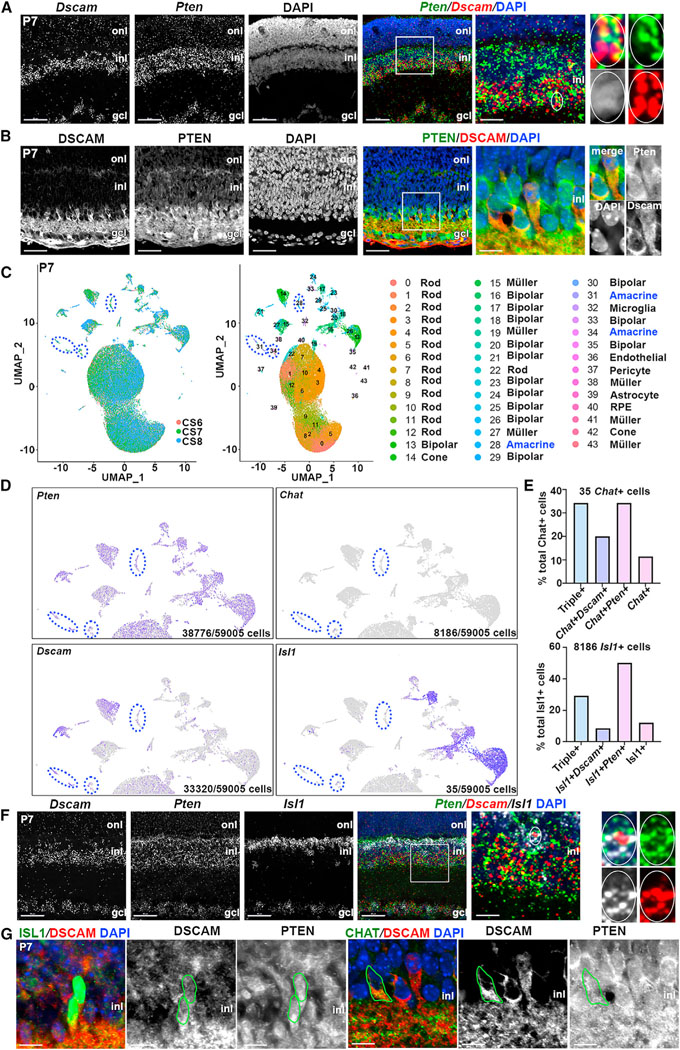
(A and B) RNAscope hybridization (A) and immunolabeling (B) of P7 retinas. High-magnification images on the right show colocalization of Dscam and Pten transcripts and proteins.
(C–E) UMAP of P7 retinal scRNA-seq data (N = 3), each sample indicated by a color code, and cluster identities (C). Pten, Dscam, Isl1, and Chat transcript distribution, with amacrine cell clusters circled in blue (D). Quantification of Chat+ and Isl1+ cells co-expressing Pten and Dscam (E).
(F) RNAscope of P7 retinas with Dscam, Pten, and Isl1 probes. High-magnification images on the right show colocalization of Dscam, Pten, and Isl1.
(G) Immunolabeling of P14 wild-type retina with PTEN, DSCAM, ISL1, and ChAT. Green circles outline ChAT+ and ISL1+ cells co-expressing PTEN and DSCAM. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer.
Scale bars: 50 μm in the first four; 10 μm in high-magnification images. See Figure S1.
To identify retinal cells expressing Pten and Dscam, we performed single-cell RNA sequencing (scRNA-seq) on three P7 wild-type retinas. High-dimensional uniform manifold approximation and projection (UMAP) plots were generated, identifying 43 unique cellular clusters (Figure 1C). Using signature gene sets for cluster annotation (Figure S1A), three amacrine cell populations were identified (Figure 1C), all of which expressed Pten and Dscam (Figure 1D). Of the 59,005 cells sequenced, only 6 expressed Th, and 35 expressed Chat, reflecting their overall rarity (<0.01% and 0.5% of all retinal cells, respectively).4,35 Further contributing to transcript scarcity, Th and Chat are mature dopaminergic and cholinergic markers, respectively, and are only beginning to be expressed by P7 (Figure S1B). To further study cholinergic amacrine cells, we examined Isl1, an early marker of bipolar cells and ChAT+ amacrine cells.36 Of the sequenced cells, 13.8% were Isl1+, of which 65% were Vsx2+ bipolar cells, and 35% were Vsx2− amacrine cells. Within the Chat+ and Isl1+ amacrine cell pools, 34% and 29% co-expressed Pten and Dscam, respectively (Figure 1E). These data were validated in situ, with Pten and Dscam co-expression detected in a subset of Chat+ and Isl1+ amacrine cells at the transcript (Figure 1F) and protein (Figure 1G) levels. Thus, Pten and Dscam are co-expressed in ~30% of cholinergic SACs at P7.
Amacrine cell radial positioning defects are exacerbated in PtencKO;Dscamdel17 double knockouts
To understand how the loss of an intracellular phosphatase (Pten) and a CAM (Dscam) similarly impact amacrine cell spacing, we generated double mutants to test epistasis. We used Dscamdel17 mice, a null allele with a 38-base pair deletion in exon 17 that creates a frameshift mutation with a premature stop codon.34 Pten conditional knockouts (cKO) were generated using a Ptenfl/fl allele with floxed exons 4 and 537 and a Pax6:Cre driver line.38 Pax6::Cre promotes recombination from embryonic day 10.5 (E10.5) in retinal progenitor cells, except in a central wedge, and, hence, all progeny neurons, including amacrine cells, are mutated in peripheral domains.38 In 56 litters collected at P14 from Ptenfl/+;Pax6::Cre+;Dscamdel17/+ with Ptenfl/+;Dscamdel17/+ crosses (376 live pups), all homozygous Dscamdel17 combinations were underrepresented compared with theoretical Mendelian ratios (Figure S2A). These findings were expected, since homozygous Dscam null mutants are perinatally lethal in a C57BL/6 genetic background.39 However, Dscamdel17 lethality was not completely penetrant, allowing us to analyze surviving pups.
Radial layers form before mosaics, so we first examined the distribution of cells in the radial axis in PtencKO and Dscamdel17 single and double mutants at P14, when cell differentiation is complete (Figures 2A and S2D–S2F). DAPI staining delineated three distinct layers in P14 wild-type retinas (Figure 2A). In P14 PtencKO retinas, retinal layering was disrupted, most notably affecting the INL and GCL, with DAPI+ nuclei located in an expanded inner plexiform layer (IPL), a normally cell-free zone (Figure 2A), as reported previously.31,40–43 Similarly, in P14 Dscamdel17 retinas, INL and GCL cells were less tightly packed, although they remained as distinct layers, and some DAPI+ nuclei were also detected in the IPL, resembling the PtencKO phenotype (Figure 2A). Finally, in P14 PtencKO;Dscamdel17 double-mutant retinas, the IPL was grossly expanded, and a “cell-free” IPL was even less distinct (Figures 2A and 2B). To quantify disturbances, we measured retinal thickness, revealing 2- and 2.2-fold increases in P14 PtencKO and Dscamdel17 retinas, respectively, and an additive 3.6-fold increase in PtencKO; Dscamdel17 retinas (Figure 2B). This expansion phenotype developed over time, since, at P7, there were 1.2- and 1.4-fold expansions in PtencKO and Dscamdel17 retinas, respectively, and a similar 1.4-fold increase in PtencKO;Dscamdel17 retinas (Figures S2B and S2C).
Figure 2. Aberrant cholinergic amacrine cell mosaics in Pten and Dscam single and double mutant retinas.
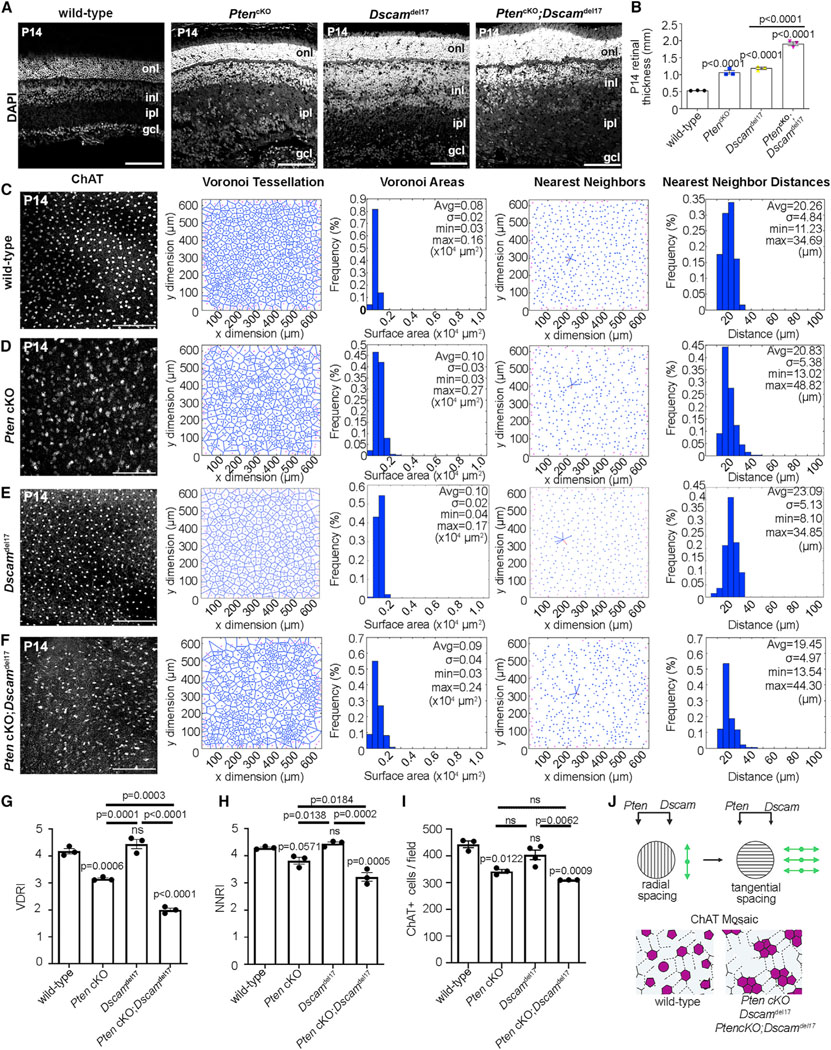
(A and B) DAPI staining of P14 wild-type, PtencKO, Dscamdel17, and PtencKO;Dscamdel17 retinal sections (A) and quantification of retinal thickness (B). Plot shows means ± SEM. N = 3 biological replicates per genotype, each with 3 technical replicates. The p values were calculated with one way ANOVA and post hoc Tukey test.
(C–H) ChAT immunolabeling and mosaic analysis of P14 wild-type (C), PtencKO (D), Dscamdel17 (E), and Pten;Dscam DKO (F) retinal flat-mounts showing Voronoi tessellations, Voronoi areas, nearest-neighbor plots, and frequency distributions for ChAT+ SACs in the INL. Shown is calculation of VDRI (G) and NNRI (H) and ChAT+ cell numbers per field.
(I) Plots show means ± SEM. N = 3 biological replicates/genotype, except N = 4 for Dscamdel17, all with 3 technical replicates. p values were calculated with one-way ANOVA and post hoc Tukey test.
(J) Summary of Pten and Dscam additive effects on amacrine cell radial and tangential migration.
Scale bars: 100 μm (A) and 200 μm (C–F). See Figures S2 and S3.
To determine whether amacrine cell radial patterning was disrupted in PtencKO;Dscamdel17 retinas, we used three markers: Pax6, a pan-amacrine cell marker that also labels some ganglion and horizontal cells;44,45 Calretinin/Calb2, which labels GABAergic amacrine cells, including the A17 subtype, and some ganglion cells;44,45 and ChAT, which labels SACs (Figures S2D–S2F). Visual inspection of all three markers confirmed the disrupted positioning of amacrine (and other) cell somata and dendritic processes in P14 PtencKO, Dscamdel17 and PtencKO; Dscamdel17 retinas (Figures S2D–S2F). Within the IPL, amacrine cells elaborate dendritic arbors that stratify to form five discrete sublaminae (s1–s5).1,46 Calretinin+ amacrine cells arborized in three IPL sublaminae (s1–s3) in P14 wild-type retinas (Figure S2E), but not in P14 PtencKO and Dscamdel17 retinas, in which only a shadow of the three layers was detected, while no stratification was visible in P14 PtencKO;Dscamdel17 retinas (Figure S2E). Similarly, SAC dendrites, which normally arborize in s2 and s4, were partially disrupted in P14 PtencKO and Dscamdel17 single-mutant retinas, and no ChAT+ dendritic stratification was visible in PtencKO;Dscamdel17 retinas (Figure S2F). As the dendritic defects were more striking when both genes were mutated, Pten and Dscam act additively, indicative of parallel pathways, to control amacrine cell radial patterning.
Pten and Dscam act additively to establish cholinergic amacrine cell mosaics
We next asked whether the loss of Pten and Dscam impacted SAC mosaic organization in the tangential plane. We began with a thorough characterization of PtencKO retinas, in which cholinergic arrays had not been assessed. Presumptive SACs were labeled with ISL1 at P736 and with ChAT at P14 and P21. Two spatial statistical measures were used to assess SAC cellular order, Voronoi domain and nearest-neighbor analyses, each derived from a Delauney tessellation of the field,47 as implemented previously.31 The frequency of Voronoi domain area sizes (being the territories closer to each cell than to any of their neighbors) and the nearest-neighbor distances were plotted, and variability was assessed using a regularity index (RI). The central retina, where Pten is not deleted by the Pax6::Cre driver, was avoided, as demarcated by a visible dorsomedial hamartoma.42
In P7 wild-type retinas, ISL1+ amacrine cell somata in the INL were distributed in a regular, patterned array, indicated by the limited variability of Voronoi domain areas (Figure S3A).6 In P7 PtencKO retinas, the Voronoi domain RI (VDRI) for ISL1+ SACs showed a 1.4-fold reduction, indicative of less regular spacing (Figures S3A and S3B). A comparable 1.2-fold reduction was observed in the nearest neighbor RI (NNRI) of ISL1+ SACs in P7 PtencKO retinas (Figures S3A and S3B). Similarly, at P14, the ChAT+ SAC VDRI was reduced 1.3-fold in PtencKO compared with wild-type retinas, while the ChAT+ SAC NNRI was reduced 1.1-fold in P14 PtencKO retinas compared with the wild type, nearly reaching significance (Figures 2C, 2D, 2G, and 2H). At P21, there was a 1.7-fold reduction in the VDRI and a 1.5-fold reduction in the NNRI (Figures S3C and S3D). SAC spatial organization is thus disrupted in the tangential plane in PtencKO retinas beginning at P7.
It has been reported that Dscamdel17 retinas have normal cholinergic arrays,48 and indeed, no differences were found between P14 Dscamdel17 and wild-type controls for either the VDRI or NNRI (Figures 2C–2H). However, the ChAT+ SAC VDRI was reduced 2.1-fold in P14 PtencKO;Dscamdel17 double mutants compared with wild-type and 1.6-fold compared with PtencKO retinas (Figures 2C–2H). The NNRI for ChAT+ SACs in P14 PtencKO; Dscamdel17 retinas was reduced 1.3-fold compared with the wild type and 1.6-fold compared with PtencKO (Figures 2C–2H). Finally, SAC density was reduced in both PtencKO and PtencKO; Dscamdel17 retinas compared with the wild type (Figure 2I). Collectively, these data suggest that Pten and Dscam act additively to regulate the spatial organization of ChAT+ amacrine cells in the radial and tangential plane (Figure 2J).
Pten regulates DSCAM localization in dopaminergic amacrine cells
To determine how PTEN, an intracellular phosphatase, and CAMs, which decorate the plasma membrane, together control amacrine cell spacing, we focused on endocytic recycling as a potential link. Newly translated CAMs are transported to the cell surface and then removed and recycled through the endocytic pathway.49 To examine whether CAM expression was disrupted in PtencKO amacrine cells, we first assessed DSCAM expression in TH+ cells since their somal mosaics and associated dendritic nets are similarly disrupted in PtencKO31 and Dscamdel17 27,34,48,50,51 retinas, albeit with more severe TH+ dendritic fasciculation defects observed upon Dscam loss (Figure S4A). DSCAM is expressed on the cell membrane and in adherens junctions, conferring a punctate distribution to DSCAM from P2 to adulthood.52 In P21 wild-type retinas, over 98% of TH+ amacrine cells had 1–3 DSCAM+ puncta, with only a few cells (1.44%) having more than 3 puncta (Figures S4B and S4C). In contrast, in P21 PtencKO retinas, over 40% of TH+ amacrine cells contained more than 3 DSCAM+ puncta, with up to 7–9 puncta per cell, which was never observed in wild-type retinas (Figures S4B and S4C). In P21 PtencKO retinas, there was a clear shift to higher numbers of DSCAM+ puncta per TH+ amacrine cell (Figure S4C).
To determine whether DSCAM+ puncta localized to specific subcellular organelle(s), we performed triple immunolabeling of TH and DSCAM with organelle markers, including CD63 to mark late endosomes/lysosomes, γ-tubulin to mark centrosomes, acetylated tubulin to mark the primary cilium, and cholera toxin β to mark lipid rafts (Figures S4D–S4G). Of these markers, DSCAM puncta only co-localized with the late endosomal-lysosomal marker CD63 (Figure S4D). Thus, Pten is required to control the balance of DSCAM protein sequestered into endosomes in TH+ amacrine cells, which may alter the normal recycling of DSCAM to the cell membrane.
Pten regulates endocytic trafficking of CAMs in amacrine cells
We turned back to SACs and analyzed the expression of two additional CAMs implicated in radial and tangential spacing: FAT35 and MEGF10.29 To identify vesicular compartments to which these CAMs localized, CAMs were co-labeled with CD63, enriched in late endosomes and lysosomes; RAB4, found in recycling endosomes; and RAB5, an early and trafficking endosome marker that co-localizes with PTEN.33 In P14 ChAT+ SACs, FAT3 and MEGF10 had punctate patterns and co-localized with CD63 (Figures 3A–3C), RAB4 (Figures 3D–3F), and RAB5 (Figures 3G–3I), suggesting that these CAMs are present in all endosomal compartments. To determine whether Pten loss influenced CAM endocytic trafficking, we compared the distribution of MEGF10 and FAT3 in P14 wild-type and PtencKO retinas. MEGF10 (Figures 3J–3L) and FAT3 (Figures 3M–3O) were expressed in the INL and IPL in a punctate pattern, including in SAC somata, but their expression was disorganized in dendritic processes throughout the expanded IPL in P14 PtencKO retinas. Furthermore, both MEGF10+ (Figure 3L) and FAT3+ (Figure 3O) puncta were increased in individual P14 PtencKO SACs.
Figure 3. Co-localization of MEGF10 and FAT3 with ChAT is disrupted in PtencKO retinas.
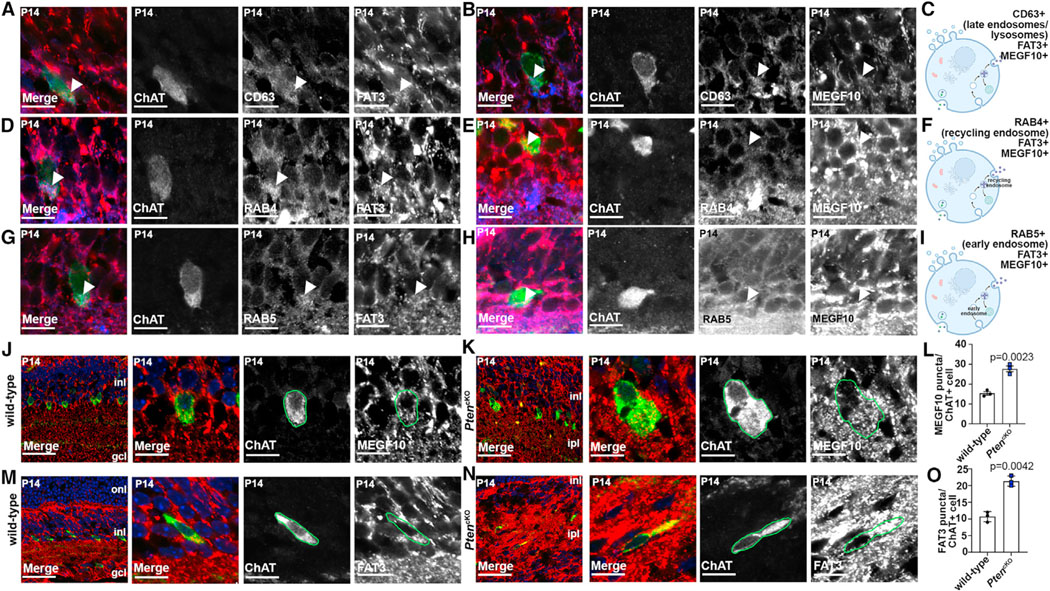
(A–C) Colabeling of P14 wild-type retinas with CD63 (blue), ChAT (green), FAT3 (red, A), or MEGF10 (red, B). Arrows point to CD63 puncta that are positive for FAT3 or MEGF10. Also shown is a schematic of CD63+ endosomal compartments (C).
(D–F) Colabeling of P14 wild-type retinas with RAB4 (blue), ChAT (green), FAT3 (red, D), and MEGF10 (red, E). Arrows point to RAB4 puncta that are positive for FAT3 or MEGF10. Also shown is a schematic of RAB4+ endosomal compartments (F).
(G–I) Colabeling of P14 wild-type retinas with RAB5 (blue), ChAT (green), FAT3 (red, G) or MEGF10 (red, H). Arrows point to RAB5 puncta that are positive for FAT3 or MEGF10. Also shown is a schematic of RAB5+ endosomal compartments (I).
(J–O) Colabeling of P14 wild-type (J and M) and PtencKO (K and N) retinas with ChAT (green) and MEGF10 (red, J and K) or FAT3 (red, M and N). Blue is DAPI counterstain. Also shown is quantification of MEGF10+ (L) and FAT3+ (O) puncta in ChAT+ cell somata in wild-type and PtencKO retinas.
Plots show means ± SEM. N = 3 biological replicates/genotype, all with 3 technical replicates, with 50 total cells analyzed for wild-type and 48 cells for PtencKO. The p values were calculated with an unpaired t test. Scale bars: 10 μm (A–H), 40 μm (J–N in the first), and 10 μm in the remaining. See also Figure S4.
Changes in the vesicular distribution of CAMs could reflect altered endocytic recycling. To test this assumption, we developed an in vitro imaging-based assay to monitor CAM trafficking off the cell membrane and into endosomal compartments. Amacrine cells were enriched from P3 retinae using magnetic-activated cell sorting (MACS) with an antibody that recognizes neural cell adhesion molecule (NCAM)(CD57, clone Vc1.1).30 Enrichment of sorted amacrine cells was confirmed by labeling with Vc.1.1 (91.00% ± 0.88%) and Pax6 (80.67% ± 0.51%) (Figure 4A). To label cell-surface CAMs and monitor their trafficking, biotin-conjugated MEGF10, FAT3, or DSCAM antibodies were added to cultured amacrine cells. PTEN signaling was disrupted using 10 mm dipotassium bisperoxo (picolinato) oxovanadate (V), or bpV(pic), a pharmacological PTEN inhibitor, whose efficacy was validated by showing an increase in pAKTSer473 levels, as expected upon PTEN inhibition (Figure 4B). We further showed that bpV(pic) disrupted the radial layering of Pax6+ amacrine cells in E18.5 retinal explants cultured for 8 days in vitro (DIV), phenocopying PtencKO retinas (Figure S5).
Figure 4. CAM endocytic recycling is accelerated in PtencKO retinas.
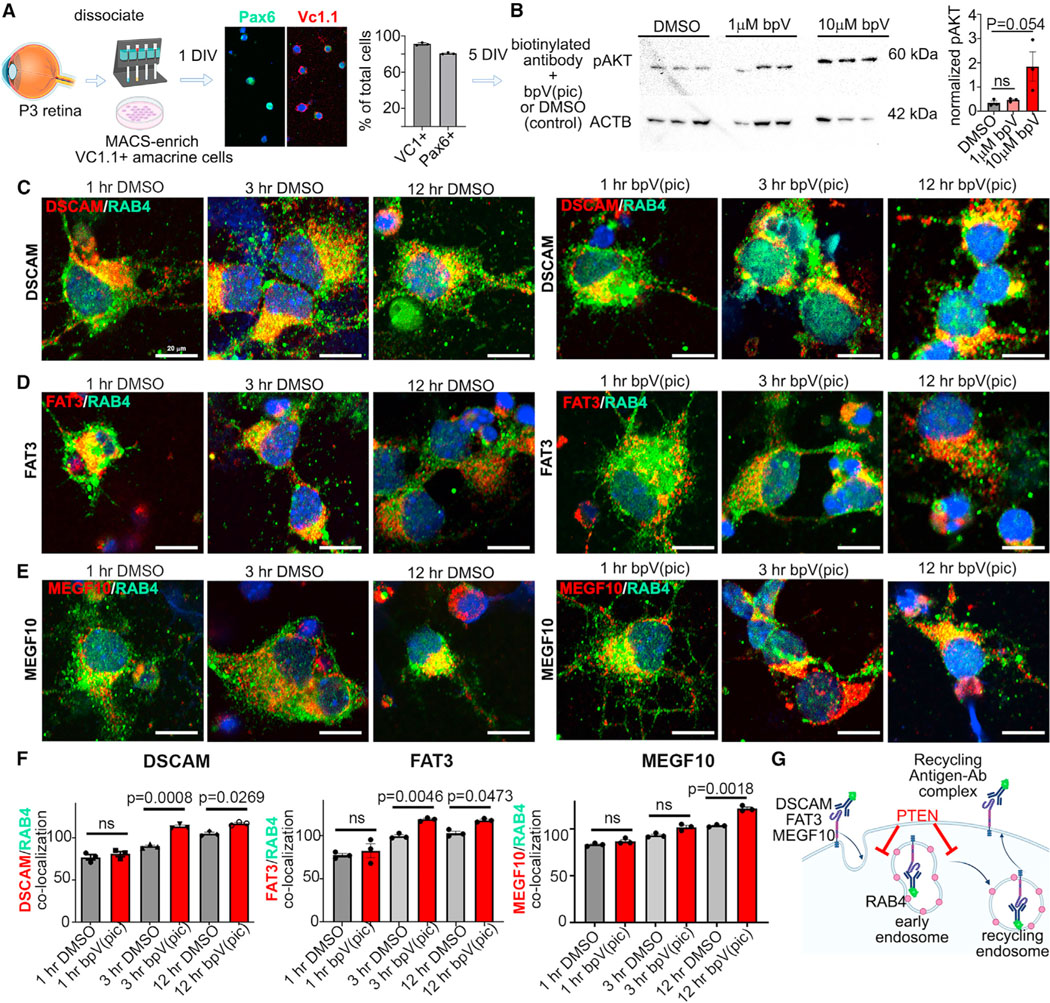
(A) Summary of the CAM endocytosis assay, showing immunolabeling and quantification of VC.1.1+ and Pax6+ MACS-enriched amacrine cells. Plots show means ± SEM. N = 3 biological replicates per sample from one experiment.
(B) Western blot of pAktSer473 in P7 retinal explants treated for 1 day with 1 μm or 10 μm bpV(pic). Densitometry is normalized to actin beta (ACTB). The plot shows means ± SEM. N = 3 biological replicates per sample from one experiment. p values were calculated with an unpaired t test.
(C–E) Immunolabeling of RAB4 with secondary antibodies to biotin-conjugated DSCAM (C), FAT3 (D), or MEGF10 (E) in MACS-enriched amacrine cells treated with DMSO or bpV(pic) for 1 h, 3 h, and 12 h.
(F) Quantification of the overlapping relative fluorescence signals of RAB4 with DSCAM, MEGF10, and FAT3. Plots show means ± SEM. 27–37 individual cells collected from 3 biological replicates were analyzed per time point and condition in 3 independent experiments. The p values were calculated with an unpaired t test in pairwise comparisons at each time point.
(G) Schematic highlighting how Pten affects endocytic trafficking.
Scale bars: 20 μm (C–E). See Figure S5.
After 1 h (baseline), 3 h, and 12 h in culture, cells were fixed and labeled with the recycling endosome marker RAB4 and with secondary antibodies recognizing biotinylated CAM anti-body-antigen conjugates (Figures 4C–4E). Internalized CAMs were evaluated by measuring overlapping fluorescence intensity between the CAM and intracellular RAB4. CAM internalization was at similar baseline levels at 1 h in both DMSO- or bpV(pic)-treated cultures (Figure 4F). Moreover, MEGF10, FAT3, and DSCAM internalization increased from 1 h to 3 h and from 3 h to 12 h, regardless of DMSO or bpV(pic) treatment (Figure 4F). However, bpV(pic) treatment accelerated the rate of DSCAM, FAT3, and MEGF10 internalization at 3h and 12h compared to DMSO controls (Figure 4F). Pten loss thus accelerates the trafficking of DSCAM, FAT3 and MEGF10 off the membrane and into endocytic vesicles in amacrine cells (Figure 4G).
Retinal cells produce EVs, including amacrine cells
Altered endosomal trafficking can impact EV biogenesis; EVs are derived from CD63+ multivesicular bodies (MVBs) that are trafficked to the plasma membrane, where intraluminal vesicles (ILVs) are released into the intercellular space as EVs (Figure S6A).53 Various retinal cells secrete EVs, including the retinal pigmented epithelium (RPE), Muüller glia, photoreceptors, and retinal progenitor cells, mainly as determined from in vitro collections of cell culture supernatants.54,55 To determine whether EV secretion was perturbed in PtencKO retinas in situ, we instead purified interstitial EVs from intact retinal tissue, which, we reasoned, would more closely capture the native tissue vesiculome.56 The RPE was removed from freshly dissected P14–P21 retinas, retinal tissue was minced and incubated for 1 h in DMEM, and small EVs/exosomes (i.e., <150 nm in diameter) were enriched by differential centrifugation with a final pelleting step at 100,000 × g. Using transmission electron microscopy (TEM), membrane-enclosed vesicles in the predicted size range of 50–150 nm were observed in P21 retinal EV preparations (Figure S6B). By western blot, EV-constituent proteins, including Flotillin-1 and TSG101,53 were detected in both P14 wild-type and PtencKO retinal EV preparations (Figure S6C). Finally, quantification of FM1–43-labeled EVs by nanoscale flow cytometry revealed that equal numbers of secreted EVs, normalized to the protein content of input cells, were produced by P14 wild-type and PtencKO retinas (Figure S6D).
We next asked whether MACs-enriched amacrine cells isolated from P12 wild-type retinas secrete EVs after 24 h in culture.30 Since ganglion cells die rapidly upon optic nerve transection and are not labeled by Vc1.1, they were not contaminants in our cultures. EVs were isolated from conditioned medium via differential centrifugation, immunolabeled with CD9, an EV-specific tetraspanin, and visualized by nanoscale flow cytometry, confirming the secretion of CD9+ EVs from amacrine cells (Figure S6E). Consistent with this finding, Smpd3, encoding neutral sphingomyelinase 2 (nSmase2), which hydrolyzes sphingomyelin to ceramide, the first step in EV production,57 was expressed in the INL and the GCL, closely matching the pattern of Pax6 transcript distribution, an amacrine cell marker, and overlapping with Pten, which was expressed throughout the INL (Figure S6F). Nevertheless, ChAT+ and TH+ amacrine cell mosaics appeared normal in adult Smpd3 null mutant (fro/fro) retinas58 (Figure S6G), suggesting other EV-biogenesis pathways, such as endosomal sorting complex required for transport (ESCRT)-dependent biogenesis, may compensate for the loss of Smpd3. Amacrine cells thus produce EVs, likely via nSmase2-dependent and -independent pathways, and Pten loss does not impact EV biogenesis.
Pten loss alters the retinal EV proteome, impacting vesicle transport proteins
Even though EV numbers did not differ between wild-type and PtencKO retinas, and Smpd3 is not required for amacrine cell patterning, EV cargo loading is fundamental to membrane trafficking. Macromolecules are loaded into EVs either passively or via cargo-initiated routes that are poorly understood.59 Due to passive loading of membrane-bound and cytosolic proteins, the vesicular proteome provides a “fingerprint” of cell status even without assigning a bioactive function to EVs. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to profile the proteomes of P14 wild-type and PtencKO retinal EVs (N = 3 retinas each; Figure 5A). 280 proteins were detected, of which 159 were in wild-type and PtencKO mutant retinas, 46 were unique to wild-type retinas, and 75 were unique to PtencKO retinal EVs (Figure 5B; Table S1). Total proteins captured were 6.5-fold lower than reported from cultured retinal progenitor cells,54 likely since extracting interstitial EVs from whole tissues remains a challenging endeavor.56 Nevertheless, several proteins involved in EV biogenesis or structure were identified in P14 retinal EVs and enriched in PtencKO retinas, including RAB10 and RAB3A, members of the Rab family of guanosine triphosphate hydrolases (GTPases), and EV surface proteins (e.g., SCAMP1, BSG, DNML, and EEF1A1)60 (Figure 5C; Tables S1 and S2). Amacrine cell-enriched proteins, including transporters (solute carriers SLC6A1, SLC6A13, SLC6A11, and SLC12A5)45 and SYNTAXIN, were also identified in retinal EVs, albeit at lower relative levels in PtencKO samples (Figure 5D; Tables S1 and S2), consistent with the overall reduction in amacrine cell numbers in mutant retinas.43 Thus, at least a subset of interstitial retinal vesicles are amacrine cell derived and retain a cell-type-specific signature.
Figure 5. The proteome of the retinal vesiculome reveals an enrichment of endocytic trafficking proteins in PtencKO retinal cells.
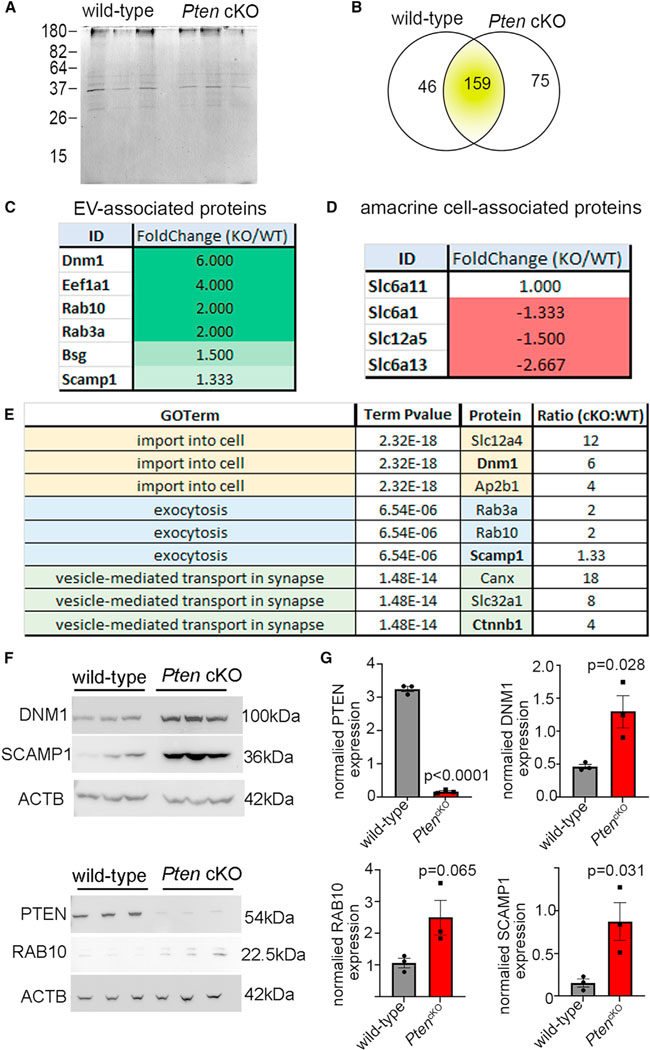
(A) Silver-stained SDS-PAGE gel with EV lysates purified from P14 wild-type and PtencKO retinas.
(B) LC-MS/MS analyses of EV proteins isolated from P14 wild-type and PtencKO retinas, showing the numbers of proteins enriched in wild-type and PtencKO samples.
(C and D) EV-associated proteins (C) and amacrine cell markers (D), showing fold change, comparing PtencKO/wild-type retinas.
(E) Biological process GO terms associated with enriched proteins in PtencKO retinal EVs.
(F and G) Western blots for PTEN, RAB10, DNM1, and SCAMP1 in P14 wild-type and PtencKO retinas. Plots show mean densitometry values normalized to ACTB ± SEM. N = 3 biological replicates/genotype.
The p values were calculated with unpaired t test. See also Figure S6.
DSCAM and MEGF10 were not detected in the proteomic datasets, likely due to the difficulty of extracting interstitial EVs, the low abundance of retinal cells expressing these proteins, and the difficulty in isolating membrane proteins for LC-MS/MS. We therefore performed western blots on P14 wild-type and PtencKO retinal EVs as a more sensitive detection method. While the absolute amount of MEGF10 was reduced in PtencKO retinal EVs, when normalized to the vesicular marker TSG101, MEGF10 vesicular content did not differ compared with wild-type retinas (Figure S6C). DSCAM was also detected in P14 retinal EVs, but levels were not different in P14 wild-type and PtencKO retinas, although the size of DSCAM (222 kDa) may have hampered accurate quantification (Figure S6H). Nevertheless, these data indicate that CAMs are detected in retinal EVs, although their relative quantities were not grossly changed in the absence of Pten.
To compare vesicular proteomes from P14 wild-type and PtencKO retinas, we performed Gene Ontology (GO) analyses of biological processes, cellular components, and molecular functions (Table S3). Specific biological process GO terms that were enriched in P14 PtencKO retinal EVs included “regulation of exocytosis,” “vesicle-mediated transport in synapse,” “plasma membrane repair,” “establishment of protein localization to plasma membrane,” and “membrane assembly” (Figures 5E and 6A; Table S4). To ascertain whether changes in the PtencKO vesiculome were indeed reflective of an altered cellular signature, we performed western blots on total tissue lysates. A gross reduction of PTEN in P14 PtencKO retinas was observed, while, conversely, several vesicular and transport-related proteins were increased in total cellular lysates, including RAB10, SCAMP1, and DNM1 (Figures 5F and 5G). Since vesicular proteomes provide a molecular fingerprint of the cellular state, these data are suggestive of a deregulation of endocytic trafficking in PtencKO retinas.
Figure 6. Canonical Wnt signaling is negatively regulated by Pten and active in migrating amacrine cells.
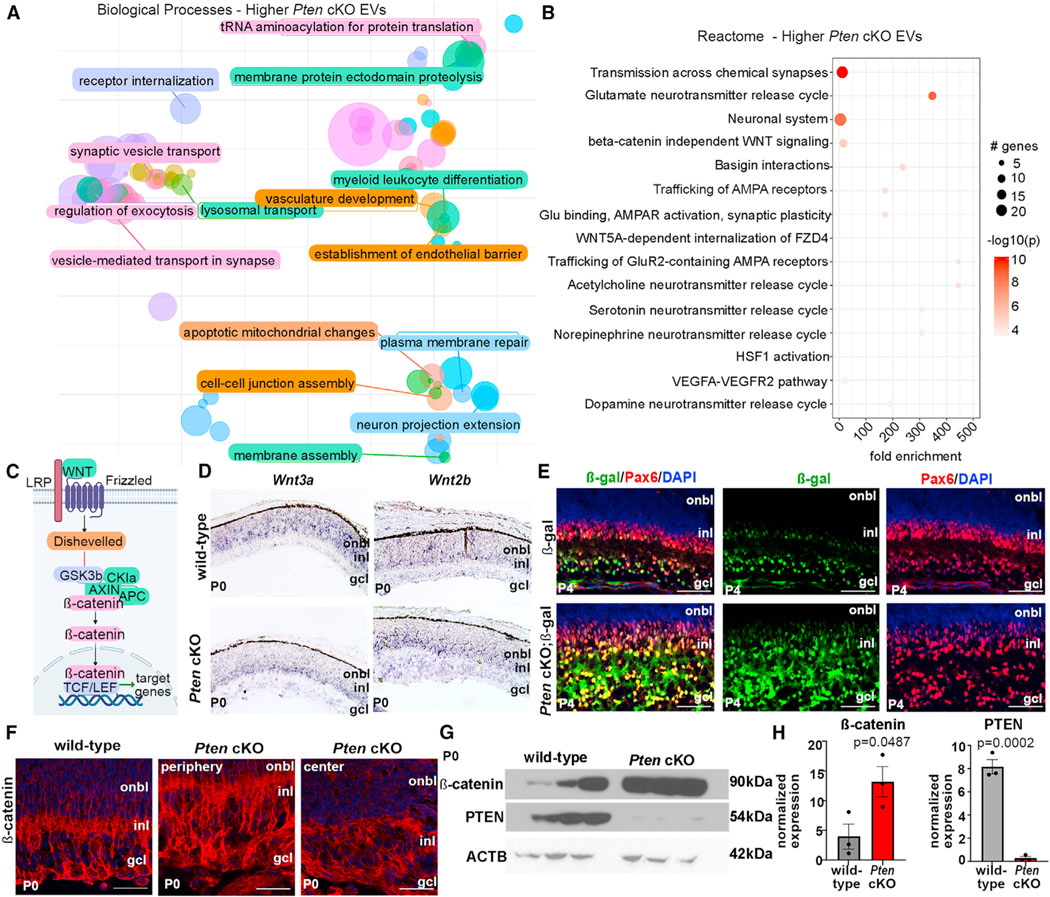
(A) Biological processes enriched in PtencKO retinal EVs.
(B) Reactome pathway analysis of enriched proteins in PtencKO retinal EVs. The size of the circle represents the number of genes enriched in the pathway; the color of the circle represents the log10 p value.
(C) Schematic of the canonical Wnt signaling pathway.
(D) Expression of Wnt3a and Wnt2b in P0 wild-type and PtencKO retinas using RNA in situ hybridization.
(E) Expression of β-galactosidase (β-gal) and Pax6 in P4 lef/tcf::lacz and PtencKO; lef/tcf::lacz retinas.
(F) Expression of β-catenin in P10 wild-type and PtencKO retinas, including in the peripheral (region of Pten deletion) and central (Pten expression is retained) retina.
(G and H) Western blots of β-catenin and PTEN in P0 wild-type and PtencKO retinas.
Plots show mean densitometry values normalized to ACTB ± SEM. N = 3 biological replicates/genotype. The p values were calculated with an unpaired t test. Scale bars: 100 μm (E and F).
Pten regulates Wnt signaling to control the spatial distribution of ChAT+ SACs
To identify top-level pathways impacted by Pten loss, we analyzed Reactome signaling pathways enriched in P14 PtencKO retinal EVs, identifying several pathways related to synaptic transmission as well as Wnt signaling (e.g., “beta-catenin independent Wnt signaling” and “Wnt5a-dependent internalization of Fzd4”) (Figure 6B; Table S5). Canonical Wnts, such as Wnt3a and Wnt2b, bind to frizzled receptors to ultimately stabilize β-catenin, which translocates into the nucleus, binds lymphoid enhancer factor/T-cell factor (LEF/TCF) transcription factors, and transactivates target genes (Figure 6C). Wnt2b and Wnt3a transcripts were detected in the developing INL and GCL in P0 wild-type and PtencKO retinas, where amacrine cells migrate (Figure 6D). To determine whether Wnt signaling is activated in amacrine cells, we used a lef/tcf-lacZ transgenic reporter.61 β-Galactosidase (β-gal) immunostaining was detected in the developing inner INL and GCL of P4 wild-type retinas, where amacrine cells are located, co-localizing with Pax6, an amacrine cell marker (Figure 6E). To quantitate Wnt signaling, we monitored non-phosphorylated (i.e., active) β-catenin expression, which was detected at low levels in the IPL of P0 and P7 wild-type retinas and upregulated in PtencKO retinas by immunostaining (Figures 6F and S7A) and by western blotting (Figures 6G and 6H). Canonical Wnt signaling is thus active in migrating amacrine cells and upregulated in PtencKO retinas.
To address whether canonical Wnt signaling is essential for amacrine cell patterning, P0 retinal explants were cultured for 7 DIV, during the period of active amacrine cell migration, and incubated with either DMSO (control; Figure 7A), recombinant Wnt3a ligand (Figure 7B), or XAV939 (Figure 7C), a tankyrase inhibitor that activates the axin-GSK3β destruction complex that degrades β-catenin (i.e., a Wnt inhibitor).62 Consistent with a role of Wnt3a in regulating amacrine cell migration, the radial organization of amacrine cells was disrupted in XAV939-treated retinal explants (Figure S7B). Furthermore, the ectopic expression of Wnt3a in E18.5 retinal explants by electroporation disrupted amacrine cell positioning in the radial plane after 8 DIV (Figure S7D). Thus, the first stage of amacrine cell positioning, radial migration, is disrupted by altering Wnt signaling.
Figure 7. Perturbing Wnt signaling alters the spatial distribution of cholinergic amacrine cells.
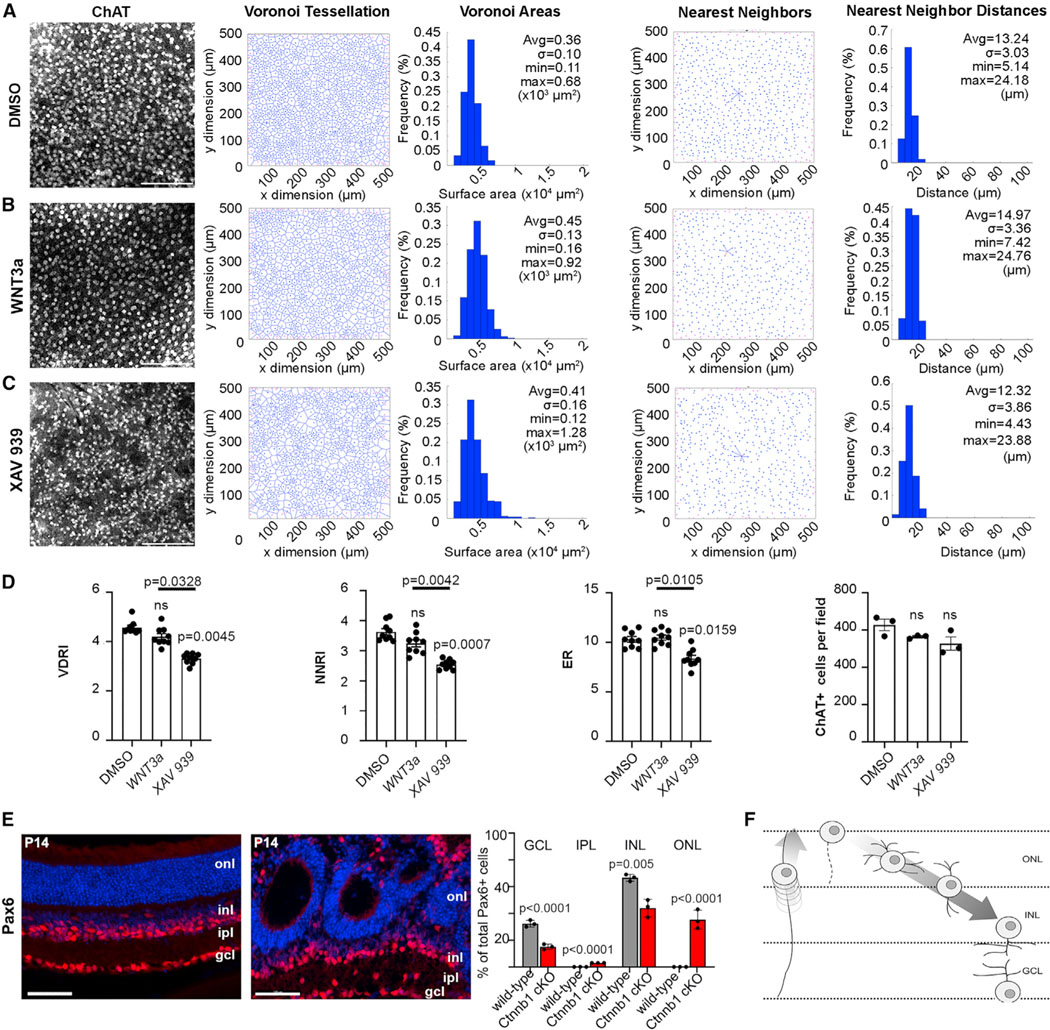
(A–C) ChAT immunolabeling and mosaic analysis of retinal explants cultured with DMSO (A), Wnt3a (B), and XAV939 (C) (N = 6 each), showing Voronoi tessellations, Voronoi areas, nearest-neighbor plots, and frequency distributions for ChAT+ SACs.
(D) Voronoi domain RI (VDRI) and nearest-neighbor RI (NNRI), effective radius (ER), and ChAT+ cell numbers in retinal explants cultured with DMSO, Wnt3a, and XAV939. Plots show means ± SEM. N = 3 biological replicates, each with 3 technical replicates in one experiment. The p values were calculated with one-way ANOVA and post hoc Tukey test.
(E) Pax6 immunolabeling and quantification of P14 wild-type and Ctnnb1cKO retinas, showing ectopic Pax6+ cells in the IPL and ONL in Ctnnb1cKO retinas. Plots show means ± SEM. N = 3 biological replicates, each with 3 technical replicates in one experiment. The p values were calculated with an unpaired t test in pairwise comparisons.
(F) Schematic of the radial migration route of newborn amacrine cells. Scale bars: 50 μm (A–C) and 100 μm (F). See also Figure S7.
After 7 DIV, the spatial distribution of SACs was assessed using Voronoi domain and nearest-neighbor analyses in retinal flatmounts. Under all conditions, SACs were densely packed (Figures 7A–7C), not respecting the ~3-diameter exclusion zones seen in wild-type retinas, presumably due to reduced retinal expansion in vitro. In explants cultured with XAV939 (Figure 7C), SACs appeared less regularly distributed relative to DMSO (control) and Wnt3a-treated explants (Figure 7A), verified by VDRI and NNRI reductions (Figure 7D). The effective radius (ER), which indicates the territory surrounding each cell where no other like-type cells can be found, averaged across the entire population of cells, was also significantly reduced in XAV939-treated explants compared with DMSO control and Wnt3a-treated explants (Figure 7D), indicating that many cells in this condition are positioned a little closer to one another than under the other two conditions. Occasional gaps in the mosaic in XAV939-treated explants were a further indication of patterning defects (Figure 7C), which arose without impacting the overall number of SACs (Figure 7D). In contrast, bath application of Wnt3a had no effect (Figure 7D), even though focal Wnt3a overexpression disrupted amacrine cell migration (Figure S7D).
To further study Wnt signaling, we crossed a floxed allele of the essential downstream effector Ctnnb1 with a Pax6-Cre driver, confirming β-catenin loss in P14 CtnnbcKO retinas (Figure S7C). The positioning of Pax6+, ChAT+, TFAP2A+, and PPP1R17+ amacrine cells was disorganized in the radial plane in P14 CtnnbcKO retinas, with fewer cells detected in the INL and GCL and ectopic amacrine cells observed in the IPL and ONL (Figures 7E and S7E–S7G). The severity of CtnnbcKO radial patterning defects, in which Wnt signaling was perturbed from the onset of retinal cell differentiation, prevented us from studying mosaics. Nevertheless, these studies confirmed the importance of Wnt signaling in guiding amacrine cell radial movements (Figure 7F). Thus, the full extent of the aberrant amacrine cell distribution in PtencKO retinas likely arises both from altered Wnt signaling and the aberrant endocytic recycling of CAMs.
DISCUSSION
In 1942, Waddington proposed the concept of developmental canalization, which suggests that built-in constraints ensure that optimal developmental programs are followed to ensure constancy of form.63 Intracellular constraints safeguard molecular functions by precisely localizing (in space and time) individual molecules to cellular membranes, the cytoskeleton, the nucleus, and/or the cytosol.64 Molecular localization is dynamic; for instance, cell surface proteins undergo endocytic recycling so that surface proteomes can meet physiological demands.65 Here, we show that Pten is a master regulator of endocytic recycling in the retina, thereby controlling the cellular distribution of CAMs and signaling molecules, to ultimately control SAC patterning. Consistent with our findings, Pten is an important regulator of endocytosis as it dephosphorylates membrane phospholipids that decorate endosomal compartments to ensure they are delivered to their appropriate destinations (e.g., recycled, exocytosed, or degraded).32 PTEN also localizes to endosomal membranes and shares sequence homology with AUXILIN, which is essential for endocytosis.66
Pten31 and Dscam34,48 regulate the radial and tangential positioning of TH+ amacrine cells and their dendritic fasciculation. From these studies, it was suggested that DSCAM is not expressed in SACs. By applying single-cell transcriptomic and RNAscope technologies, we found that Pten and Dscam transcripts are co-localized in either a distinct SAC subpopulation or in a transitory maturation phase during SAC development. Furthermore, by performing single- and double-mutant analyses, we revealed that Pten and Dscam act in parallel, partially redundant pathways to regulate tissue patterning, so that the most severe patterning disruptions are seen in Pten;Dscam double mutants. At the genetic level, these studies suggest that Pten and Dscam act additively to regulate SAC spacing. However, whereas Pten and Dscam are both essential contributors to the formation of TH+ mosaics, SAC mosaics primarily depend on Pten, with a minor contribution from Dscam, as Dscam loss affected SAC spacing only in the context of a PtencKO.
Dscam requirement to control SAC mosaic formation in the context of a PtencKO phenotype was unexpected, since single Dscam KOs do not have a phenotype (our study and Fuerst et al.48). The most parsimonious explanation is that Pten lies at the top of the amacrine cell spacing cascade by controlling endocytic recycling of multiple CAMs, each with their own independent role(s). By deleting both Pten and Dscam, we globally altered CAM processing and removed DSCAM, which acts as a “non-stick coating” to prevent cell clumping.26,48,50 Furthermore, FAT3, which is required for the multipolar → bipolar → unipolar transitions of migrating amacrine cells,67 is more quickly endocytosed in PtencKO retinas, as is MEGF10, which mediates dendrite-soma exclusion to establish SAC mosaics.10,29 Cholinergic amacrine cells that rely on Megf10 for positioning for the most part function unimpeded by Dscam.29 However, in the ~30% of ChAT+ and ISL1+ SACs that normally express Dscam, either in a short temporal window or in a subpopulation of cells, the loss of this ‘non-stick coating’ could contribute to amacrine cell clumping.
EVs were initially implicated as a waste management system to get rid of unwanted cellular components but are now viewed as an essential mode of intercellular signaling59 and are used as biomarkers of the cell state, especially in the cancer field.68 We found DSCAM and MEGF10 are in retinal EVs, and future studies will test the significance of these secreted CAMs. Interestingly, the N-terminal region of DSCAM is secreted into the medium in vitro, but potential functions in neural development have not been identified.69 The entire Wnt/β-catenin signalosome is endocytosed into MVBs in responding cells, preventing β-catenin degradation.70 Noncanonical Wnt signaling is important for retinal OPL formation,71 whereas canonical Wnt signaling is understudied. We found that Wnt signaling is activated in migrating amacrine cells, and its pharmacological perturbation phenocopies PtencKO amacrine cell patterning defects. We further found that Pten normally inhibits Wnt signaling, as expected, because in the absence of Pten, phosphatidylinositol 3-kinase (PI3K)/Akt signaling is elevated in the retina,43 and Akt inhibits GSK3, part of the β-catenin destruction complex.72 Consequently, β-catenin is stabilized, and Wnt signaling is elevated in PtencKO retinas.
DSCAM transiently co-localizes with synaptic markers in retinal puncta between P10 and P14,52 likely reflecting the endosomal sorting of synaptic vesicles during recycling.73 Several synaptic proteins are enriched in PtencKO EVs, suggestive of an increase in synapse formation. Indeed, syntaxin, a presynaptic marker, is upregulated in PtencKO retinas.31 The association between Pten loss and synaptic remodeling has been studied in the hippocampus, with Pten loss increasing connectivity.74 This association is of interest since Pten loss or the consequent mTOR activation is associated with autism and epilepsy, characterized by altered neuronal activity.75,76 A Pten-dependent mode of molecular constraint may be generalizable to the brain, where patterning defects are also observed upon Pten loss.77
Limitations of the study
In this study, we used a PtencKO that deletes Pten in retinal progenitor cells and their progeny and a Dscamdel17 allele that impacts all cells. To directly assess the role of Dscam and Pten in SAC pattern formation versus a general role in patterning the retina, conditional ablations of either or both genes should be performed in SACs. We also relied on proteomic signatures of interstitial EVs, which are difficult to isolate from intact tissues,56 meaning that the vesicular proteome that we identified here is likely an incomplete representation. Nevertheless, we took this approach as the best way to interrogate how EVs might participate in cell-cell communication in the intact retina. Furthermore, with this approach, we were able to identify and validate the utility of the interstitial EV proteome for fingerprinting the cell of identity, leading to the identification of Pten-regulated vesicular processes, including unappreciated links to endocytic processing and Wnt signaling. As methods improve for isolating interstitial EVs, new insights into biological events in tissue microenvironments are likely to be uncovered.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Carol Schuurmans (cschuurm@sri.utoronto.ca).
Materials availability
Transgenic mouse lines generated in this study are deposited in a central repository (Jackson Laboratory). All antibodies and reagents are commercially available.
Data and code availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD: 036221 and https://doi.org/10.6019/PXD036221. Uncropped western blot images were deposited in Mendeley data: 10.17632/jf4pwztxrj.1 (https://doi.org/10.17632/jf4pwztxrj). scRNASeq data were deposited on Gene Expression Omnibus with accession number GEO: GSE228191
This paper does not report original code
All other data reported in this study will be shared by the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
None of our experimental animals had been previously used for other procedures. All animals were healthy.
Mice
All animal procedures were approved by the University of Calgary Animal Care Committee (AC11–0053) and later by the Sunnybrook Research Institute Animal Care Committee (16–606) in agreement with the Guidelines of the Canadian Council of Animal Care (CCAC). The following animal lines were used from Jackson Laboratory: Ptenfl allele37 (B6.129S4-Ptentm1Hwu/J. Strain #: 006440, RRID:IMSR_JAX:006440), Pax6::Cre driver38 (STOCK Tg(Pax6-GFP/cre)1Rilm/J. Strain #:024578. RRID:IMSR_JAX:024578. Common Name: P0–3.9GFPCre), fro/fro,58 Dscamdel1734 B6.CBy-Dscamdel17/RwbJ. Strain #:008000. RRID:IMSR_JAX:008000) and lef/tcf-lacZ reporter mice (STOCK Tg(TCF/Lef1-lacZ)34Efu/JStrain #:004623. RRID:IMSR_JAX:004623). Common Name: TOPGAL). Ctnnb1fl: B6(Cg)-Ctnnb1tm1Knw/J. RRID:IMSR_JAX:022775). PCR genotyping was performed following Jax protocols. Lines were maintained by crossing with C57BL/6J wild-type mice (Strain #:000664. RRID:IMSR_JAX:000664. Common Name: B6). Animal explants were performed with CD1 mice (Charles River, IMSR_CRL:022). The day of the vaginal plug was considered to be embryonic day (E) 0.5 for timed pregnancies.
METHOD DETAILS
Tissue processing
Eyes were dissected out from embryos or postnatal pups at the designated stages and either fixed directly or the retinas were first dissected out. Fixation of eyes or retinas was achieved by immersing tissues in 4% paraformaldehyde (PFA)/1X phosphate buffered saline (PBS) at 4°C overnight for dissected retinas or eyes. For retinal explants, a shorter fixation time of 3 h was applied. For all tissues, post fixation we applied 3 × 10 min washes in 1X PBS before the tissues were cryopreserved in 20% sucrose/1X PBS overnight at 4°C. Cryoprotected tissues were embedded in Optimal Cutting Temperature (O.C.T) compound (Tissue-Tek, Sakura Finetek U.S.A. Inc., Torrance, CA) and frozen on dry ice. Cryosections were cut at 10 μm on a Leica CM3050s cryostat (Leica Biosystems, Buffalo Grove, IL, USA). Sections were collected on Fisherbrand Superfrost Plus slides (Thermo Fisher Scientific, Markham, ON).
RNAscope and RNA in situ hybridization
RNA in situ hybridization using digoxygenin labeled Wnt3a and Wnt2b riboprobes was performed as previously described.82,83 RNAscope in situ hybridization was performed using a RNAscope Multiplex Fluorescent Detection Kit v2 (ACD; #323110) according to the manufacturer’s directions. ACD probes included: Mm-Pten (#316301-C1), Mm-Dscam (#834031-C2), Mm-Th (#317621-C3), Mm-Isl1 (#451931-C3) and Mm-Smpd3 (#815591-C3). Opal 520 (Akoya #FP1487001KT; 1:1500), Opal 570 (Akoya #FP1488001KT; 1:1500) and OpalTM 690 (Akyoya #FP1497001KT; 1:1500) were used to stain channel 1, 2 and 3 probes. Retinal sections were counterstained with DAPI and mounted in Aqua-Poly/Mount (Polysciences #18606–20) as described by the manufacturer.
Immunohistochemistry
Section31,43 and flatmount31 immunofluorescence was performed as previously described. The following primary antibodies were used: Acetylated-tubulin (1:500, Abcam #ab11323), TFAP2A (Abcam#ab52222), β-catenin (1:500, H-102, Santa Cruz #sc7199), β-galactosidase (1:400, Chemicon #AB986), Calretinin (1:2000, Swant #76699/4), CD63 (1:200, BD Pharmingen #556019), ChAT (1:500, Chemicon #AB144P), Cholera Toxin β (1:500, Invitrogen/Life #C-34776), DSCAM (1:100, R&D Systems #AF3315), DSCAM (1:200, R&D Systems #AF3666), DSCAM (Bioss, bs-11456R-Biotin), FAT3 (1:100, Santa Cruz #sc-133366), FAT3 (Biotin) FAT3 (Antibodies, Online, ABIN6882377), Flotillin-1 (1:500, Cell Signaling #3253), γ-tubulin (1:500, Sigma #T3195), GFP (1:500, Abcam #ab5450), MEGF10 (1:500, Millipore #ABC10), MEGF10 Biotin conjugated (Thermo Fisher Scientific, BS-24335R-BIOTIN), PAX6 (1:500; Covance Research #PRB-278P), PPP1R17 (Atlas antibodies# HPA047819), PTEN (1:500, Cell Signaling #9559), RAB4 (1:500, Novus Biologicals #NBP2–37845), RAB5A (1/500, Proteintech #66339–1-Ig), TH (1:500, Millipore #AB152), and Vc.1.1 (Anti-HNK-1/N-CAM (CD57), 1:500, Sigma-Aldrich, C6680). Slides/tissues were washed 3 times in PBS with 0.1% Triton X-100 (PBT) and primary antibodies were detected using secondary antibodies conjugated with Alexa 568 (1:500, Molecular Probes), Alexa 488 (1:500, Molecular Probes), Alexa 647 (1:500, Molecular Probes), and 1:500 Streptavidin Cy3 (Thermo Fisher Scientific, #434315). Retinal sections were counterstained with DAPI and mounted in Aqua-Poly/Mount.
Amacrine cell sorting and culture
Retinas dissected from P3 CD1 pups were dissociated into single cells using a Papain dissociation kit (LK003150, Worthington Biochemical Corp) following the manufacturer’s instructions. Amacrine cells were purified by magnetic activated cell sorting (MACS) using a VC.1.1 antibody. Briefly, dissociated single cells were resuspended in MACS buffer (PBS/BSA 0.5%/EDTA 2mM), incubated for 30 min with VC.1.1 antibody (400ng Ab/107 cells) on ice, then washed 3 times in MACS buffer. Cells were then incubated with anti-mouse micro beads (Miltenyi Biotec, #130–047-302) for 30 min on ice and washed 3 times with MACS buffer. MS columns were used to perform magnetic separation. VC1.1 positive cells were collected washed and cultured in 8 chamber culture slides pre-coated with Poly-D-Lysine (10 mg/mL) (Corning, 354210)/Laminin (Corning, 354232) (2.5 μg/mL) in amacrine cell media: Neurobasal (Invitrogen 21103–049), B27 (1X) (Invitrogen, 17502–048), N2 (1X) (Invitrogen, 17502048), NAC (Sigma cat # A8199), Insulin (5 ng/ml) (Invitrogen, 12585–014), Na Pyruvate (1mM) (Invitrogen 11360–070), L-glutamine (2mM) (Invitrogen 25030–081), T3 (50 ng/ml) (Sigma T6397), Pen/Strep (1X) (Wisent bioproducts 450–200-EL). Amacrine cell media was supplemented immediately before plating with: BDNF 50 ng/ml (Peprotech, 450–02), CNTF 10 ng/ml (Peprotech, 450–13), TGF-β1 5 ng/ml (Peprotech, 100–21) & TGF-β2 5 ng/ml (Peprotech, 100–35B), Forskolin (1X) (Sigma, F6886).
Endocytosis assay
VC1.1+ amacrine cells were cultured for 5 DIV in presence of 1/500 biotin-conjugated DSCAM (Bioss, #bs-11456R-Biotin), FAT3 (Biorbyt, # orb400315) and MEGF10 (Thermo Fisher Scientific, BS-24335R-BIOTIN) antibodies. Additionally, DMSO (control) or 10 μm bpV(pic) (Selleckchem, S8651) were added to different wells. At the experimental time points (1hr, 3hr, 12hr), cells were washed and fixed in 4% PFA for 15min and incubated with Streptavidin-Cy3 for 30min. The cells were also immunostained with RAB4 antibody (Novus Biologicals, NBP2–37485) and a secondary antibody. Nuclei were stained with DAPI and coverslips were mounted in Aqua-Poly/Mount. To measure antibody internalization, CAM staining that co-localized with RAB4 was selected by channel overlap and the fluorescence intensity was measured using ImageJ.78
EV purification
For EV purification, eyes were enucleated, the RPE, cornea, lens and blood vessels were removed, and retinas were collected and briefly rinsed with PBS. The retina was placed on a dry 10 cm Petri dish, and minced into smaller pieces using a scalpel. The minced tissues were then transferred into a 15 mL corning tube with 10 mL pre-warmed DMEM. The retinal tissues were further dissociated by triturating with a 5 mL pipet. Dissociated retinal cells were incubated in a 37°C water bath for 1 h with gentle rocking, allowing cells to secrete EVs into the medium. Following the incubation, EVs were isolated using differential centrifugation as previously described84 with the following speeds (300 x g, 5 min; 3024 x g, 15 min; 15,475 x g, 15 min; 153,297 x g, 75 min; 153,297 x g, 75 min). EV pellets were resuspended in 50 μL of cold PBS with 1 x protease inhibitor and 1 x sodium azide and were then further processed for EM imaging or western blotting experiments.
Transmission electron microscopy
4 μL of purified exosomes were placed on carbon-coated Cu400 TEM grids that were first glow discharged for 30 s (Pelco EasiGlow, Ted Pella Inc.). After 1 min, excess liquid was wicked away, and then washed with 4 μL of distilled water 3 times. 4 μL of 2% uranyl acetate solution was applied for staining for 30 s, with excess liquid wicked away before the grids were air-dried. Grids were imaged on a Thermo Fisher Scientific Talos L120C TEM operated at 120 kV using a LaB6 filament in the Microscopy Imaging Laboratory, University of Toronto.
Nano-flow cytometry
Amacrine cells were isolated from dissociated P12 retinas using MACS. 1 μL of purified EVs were diluted in 18 μL H2O and then labeled with 1 μL anti-CD9 (Santa Cruz; #sc9148) for 30 min at room temperature. EVs were washed in sterile water, and then stained with Alexa Fluor 647 secondary antibody (Invitrogen, 1:20 dilution) for 20 min. EVs were washed in sterile water and resuspended in 500 μL sterile water and quantified on the Nanoscale Flow Cytometer (Apogee Flow Systems Inc). FM1–43 dye (Invitrogen; cat #T3163) was used to detect and measure the EV levels using nanoscale flow cytometry. To do so, 5uL sample was mixed with 2uL dye and incubated for 10 min in 37°C water bath. The mixture was then diluted by adding 500uL PBS and total FM1–43+ EV particles were measured using Apogee MicroPLUS Flow Cytometry. The results were normalized to the number of cells. EV number is quantified from scatterplots of Alexa 647 emission (CD9+ levels) and long angle light scatter (LALS), for size distribution). EVs were defined as particles greater than 100 nm in diameter.
Western blotting and silver staining
Retinas were lysed in NP-40 lysis buffer (0.05 m Tris, pH 7.5, 0.15 m NaCl, 1% NP-40, 0.001 m EDTA) with protease (1X protease inhibitor complete, 1 mM PMSF), proteasome (MG132 at 0.05 mM) and phosphatase (50 mM NaF, 1 mM NaOV) inhibitors. 10 μg of lysate from whole cells or tissue was run on SDS-PAGE gels for western blot analysis. To characterize purified EVs, they were boiled at 95°C for 2 min to break the EV membrane. The protein concentrations were determined using a Micro BCA Protein Assay Kit (ThermoFisher Scientific, #23235) following the manufacturer’s instructions. 2 μg of lysate was incubated with the blots overnight at 4°C. Primary antibodies included: β-actin/ACTB (1:10000, Abcam #8227), DSCAM (1:1000, ThermoFisher #PA5–42698), Flotillin-1 (1:1000, Cell Signaling #3253), MEGF10 (1:1000, Millipore #ABC10), PTEN (1:1000, Cell Signaling #9559), ß-Catenin-non-phospho (Active) (Ser33/37/Thr41) (D13A1) (1:1000; Cell Signaling #8814), pAKTser473 (1:1000; Cell Signaling #4060), SCAMP1 (1:1000, SYSY #121002), RAB10 (1:1000, Abcam #ab104859), DNM1 (1:1000, Invitrogen #PA1–660) and TSG101 (1:1000, Sigma #HPA006161). Blots were washed 3 × 10 min in TBST before incubating in secondary antibodies, including either Goat Anti-Rabbit IgG (H + L)-HRP Conjugate (1/50,000; Bio-Rad #1721019 or Cell Signaling #7074) or Goat Anti-Mouse IgG (H + L)-HRP Conjugate (1/50,000; Bio-Rad #1721011). Western blot signals were converted to a chemiluminescent signal using an Amersham ECL Prime Western Blotting Detection Reagent (Cytiva #RPN2232) according to the manufacturer’s instructions. Signal was detected by exposing to autoradiography film (LabForce #1141J52) or using a Bio-rad gel doc and ImageLab software. Each western blot was performed on lysates from three independent retinas, and densitometries were calculated using UN-SCAN-IT gel densitometry software (Silk Scientific) or ImageJ Software. Absolute values (“relative”) or normalized values (over β-actin/ACTB) were plotted as indicated.
LC-MS/MS
EVs were lysed in NP-40 lysis buffer and 2 μg of total protein was separated on a 10% SDS-PAGE gel and silver stained. Proteins were digested using trypsin as described.85 Peptide extracts were concentrated by Vacufuge (Eppendorf). Peptides were analyzed by LC-MS/MS using a Dionex Ultimate 3000 RLSC nano HPLC (Thermo Scientific) and Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific) at the Ottawa Hospital Research Institute Proteomics Core Facility (Ottawa, Canada) as described.85 MASCOT software version 2.6 (Matrix Science, UK) was used to infer peptide and protein identities from the mass spectra. The observed spectra were matched against mouse sequences from SwissProt (version 2016–09) and also against an in-house database of common contaminants. Results were then exported to Scaffold version 4.8.2 (Proteome Software, USA) for further analysis. Contamination cleanup was performed by labeling DECOY proteins, unknown, non-mouse proteins and keratin proteins, which were removed. Enriched proteins Briefly, proteins identified with less than 2 peptides and proteins with a probability of identification of less than 95% were filtered out. ClueGO, a Cytoscape plug-in,80 was used to annotate biological process GO terms that were higher in PtencKO retinal EVs. Qiagen Ingenuity Pathway Analysis (IPA) was used to visualize fold-changes in protein levels. REACTOME enriched pathway clustering was obtained using pathfind R Bioconductor package,81 extracting four types of plots: 4 types of plots: 1) Simple dot plot for top 15 pathways; 2) Clustered dot plot that shows that clusters together similar pathways; 3) Term-gene enrichment map; 4) UpSet plot that shows genes in enriched pathways. For gene ontology analysis, we present Scatterplots that reflect semantic similarities and treemaps using a rrvgo Bioconductor package https://bioconductor.org/packages/release/bioc/vignettes/rrvgo/inst/doc/rrvgo.html.
scRNA-seq
P7 wild type retinas were dissected and dissociated into single cells using a Papain kit (LK003150, Worthington Biochemical Corp) according to the manufacturer’s instructions. Dead cells were eliminated using a Dead cell removal kit (130–090-101, Miltenyi Biotec) following the manufacturer’s instructions. Only samples with viable cells percentages of 90% or greater were selected to proceed. Cells viability was determined using Trypan blue staining. Cell suspensions of 1X 10^6 cells/ml were prepared and loaded into GEM Chip K for an expected recovery of 10,000 cells using Chromium Next GEM Single Cell 5′ Reagent Kit v2 (10X genomics, Cat #1000263). Libraries were generated according to the manufacturer’s instructions using 50 ng of cDNA. Equal molar quantities of each library were sequenced using the Illumina NovaSeq S4 flow cell system (The Center for Applied Genomics, The Hospital for Sick Children, Toronto).
scRNA-seq analysis
10x scRNA-seq data was processed and analyzed using the Seurat v.3.2.3 R package.79 Low quality cells were excluded by filtering out cells with fewer than 500 detected genes and cells with mitochondrial RNA more than 5%. The data was then transformed by the SCTransform function while regressing out the variance due to mitochondrial RNAs. Clustering was performed by the RunPCA, FindNeighbors and FindClusters functions using the first 30 principal components. The 2D projection of the clustering was carried out by the RunUMAP function. The annotation of cell type to each cluster was performed by using markers in86 (Figure S1). Expression of selected genes was plotted using the FeaturePlot function.
Retinal explants
Eyes were removed from P0 CD1 pups using surgical forceps and placed in 1x PBS, pH 7.4 on ice. The eye was then held by the cornea to remove the iris, lens and anterior segment. The retina was then removed from the underlying retinal pigment epithelium (RPE), choroid and sclera. Isolated retinas were transferred to a sterile 10 cm dish in cold 1x PBS pH 7.4 for further processing. Retinas were flatmounted onto 13 mm, 0.8 μm Nuclepore Track-Etch membrane (GE Healthcare, catalog number: 110409) and floated on 2 mL of retinal explant media (50% DMEM, 25% HBSS, 25% heat-inactivated horse serum, 200 μM L-glutamine, 0.6 mM HEPES, 1% Pen-Strep) in individual wells of a 24-well plate. Recombinant Wnt3a protein (Catalog # 5036, R&D Systems, final concentration of 100 ng/mL in water), XAV939, a Wnt antagonist (Catalog #S1180, Selleckchem; final concentration of 10 mM in DMSO), or DMSO was added to the culture media. Half the media was removed and replaced with fresh media containing drugs every second day. For electroporation with pCIG2 and pCIG2-Wnt3a, 10μL of DNA (1 μg/μl) was applied to each explant, followed by 7 × 50ms pulses of 50mV with a BTX electroporator. Retinas were then cultured in retinal explant media as described.
Cell spacing analysis
Flat-mounted retinas or retinal explants were imaged through the z axis to ensure all ChAT+ and Islet+ cells were included in the images. ImageJ was used for collecting the X-Y coordinates of ChAT+ and Islet1+ amacrine cells. These values were imported into a customized program that computes the Voronoi tessellation of the field and the nearest neighbor distances of each individual cell, from which the regularity indices of the distribution of Voronoi areas and nearest neighbor distances were calculated (mean/standard deviation). Explant cultures were also assessed for minimal spacing between cells within a field, expressed as the effective radius (ER) derived from the density recovery profile.87 Border cells with uncertain spatial statistics were excluded from the measurements, all as previously described.47,88
Imaging
Digital images were captured using fluorescent light from a Leica DM18 inverted microscope and Leica Application Suite X (LASX) software for cell counts. Confocal images were captured using a Nikon A1 laser scanning confocal microscope available at the Center for Cytometry and Scanning Microscopy at Sunnybrook. Adobe Photoshop 2021 was used to make Figures. A license to BioRender.com or the software Inkscape 0.48 was used to prepare schematics.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed on a minimum of three animals per each genotype (N; biological replicates) with a minimum of three retinal sections from each animal (n; technical replicates). Individual dots in each plot correspond to a single biological replicate (N) after averaging the three technical replicates (n). All statistical analysis and graph designs were performed using GraphPad Prism Software version 8.0 (GraphPad Software). Error bars represent standard error of the mean (SEM). Unpaired two tailed Student’s t test was used to calculate statistical significance between two experimental groups. For multiple comparisons of more than two groups, a one-way ANOVA was used followed by a Tukey post hoc-test.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Mouse anti-acetylated Tubulin | Abcam | Cat# ab11323 RRID:AB_297928 |
| Mouse anti-β-Actin (loading control) | Abcam | Cat#ab8227 RRID:AB_2305186 |
| Rabbit anti-β-Catenin (H-102)- total | Santa Cruz | Cat# sc7199 RRID:AB_634603 |
| Rabbit anti-β-Catenin Non-phospho (Active)(Ser33/37/Thr41) (D13A1) | Cell Signaling Technology | Cat# 8814 RRID:AB_11127203 |
| Rabbit anti-β-Galactosidase | Chemicon (Millipore Sigma) | Cat# AB986 RRID:AB_92401 |
| Rabbit anti-Calretinin | Swant | Cat# 76699/4 RRID: N/A |
| Rabbit anti-CD9 | Santa Cruz | Cat# sc9148 RRID:AB_2075905 |
| Mouse anti-CD63 | BD Pharmingen | Cat# 556019 RRID:AB_396297 |
| Goat anti-ChAT (choline acetyltransferase) | Chemicon | Cat# AB144P RRID:AB_2079751 |
| Goat anti-human DSCAM Affinity Purified Polyclonal | R&D Systems | Cat# AF3666 RRID:AB_2230818 |
| Rabbit anti-DSCAM Polyclonal | Thermo Fisher Scientific | Cat# PA5–42698 RRID:AB_2610175 |
| Rabbit anti-DSCAM (Biotin conjugated) | Bioss | Cat# bs-11456R-Biotin RRID: N/A |
| Rabbit anti-Dynamin 1 polyclonal antibody (DNM1) | Thermo Fisher Scientific | Cat# PA1–660 RRID:AB_325845 |
| Rabbit anti-FAT3 (E—12) | Santa Cruz | Cat# sc-133366 RRID:AB_10655386 |
| Rabbit anti-FAT3 (Biotin-conjugaged) | Antibodies-Online | Cat# ABIN6882377 RRID: N/A |
| Goat anti-GFP | Abcam | Cat# ab5450 RRID:AB_304897 |
| Mouse anti-HNK-1/N-CAM (CD57) clone Vc1.1 | Sigma-Aldrich | Cat# C6680 RRID:AB_1078474 |
| Rabbit anti-MEGF10 | Millipore | Cat# ABC10 RRID:AB_11204003 |
| Rabbit anti-MEGF10 (Biotin-conjugated) | Thermo Fisher Scientific | Cat# BS-24335R-BIOTIN RRID: N/A |
| Rabbit anti-PAX6 | Covance Research | Cat# PRB-278P RRID:AB_291612 |
| Rabbit anti- Phospho-Akt (Ser473) (D9E) | Cell Signaling Technology | Cat# 4060 RRID:AB_2315049 |
| Rabbit anti-PTEN | Cell Signaling | Cat# 9559 RRID:AB_390810 |
| Mouse anti-RAB4 | Novus Biologicals | Cat# NBP2–37485 RRID: N/A |
| Mouse anti-RAB5A | Proteintech | Cat# 66339–1-IG 66339–1-IG |
| Rabbit anti-RAB10 antibody [4E2] | Abcam | Cat# Ab104859 RRID:AB_10711207 |
| Rabbit anti-SCAMP1 | Synaptic Systems | Cat# 121 002 RRID:AB_887780 |
| Rabbit anti-TH | Millipore | Cat# AB152 RRID: AB_390204 |
| Rabbit anti-TSG101 | Sigma-Aldrich | Cat# HPA006161 RRID:AB_1080408 |
| Rabbit anti-Tubulin, gamma (DQ-19) | Sigma-Aldrich | Cat# T3195 RRID:AB_261651 |
| αMouse IgG Alexa Flour 647 | Life Tech. | Cat# A-31571 RRID:AB_162542 |
| αMouse IgG Alexa Flour 555 | Life Tech. | Cat# A32773 RRID:AB_2762848 |
| αMouse IgG Alexa Flour 488 | Life Tech. | Cat# A32766 RRID:AB_2762823 |
| αGoat IgG Alexa Flour 488 | Life Tech. | Cat# A11055 RRID:AB_2534102 |
| αGoat IgG Alexa Flour 568 | Life Tech. | Cat# A11057 RRID:AB_2534104 |
| αRabbit IgG Alexa Flour 488 | Life Tech. | Cat# A-21206 RRID:AB_2535792 |
| αRabbit IgG Alexa Flour 568 | Life Tech. | Cat# A10042 RRID:AB_2534017 |
| αRabbit IgG Alexa Flour 647 | Life Tech. | Cat# A-31573 RRID:AB_2536183 |
| αRat IgG Alexa Flour 568 | Jackson | Cat# 711–156-152 RRID:AB_2340603 |
| Streptavidin Cy3 | Thermo Fisher Scientific | Cat# 434315 RRID: N/A |
| Goat Anti-Rabbit IgG (H + L)-HRP Conjugate | Bio-Rad | Cat# #1721019 RRID: AB_11125143 |
| Goat Anti-Mouse IgG (H + L)-HRP Conjugate | Bio-Rad | Cat# #1721011 RRID: AB_11125936 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Opal™ 690 reagent | Akoya | Cat# FP1497001KT RRID: N/A |
| Opal™ 570 reagent | Akoya | Cat# FP1488001KT RRID: N/A |
| Opal™ 520 reagent | Akoya | Cat# FP1487001KT RRID: N/A |
| XAV939 | Selleckchem | Cat# S1180 RRID: N/A |
| Cholera Toxin Subunit β-Alexa Fluor™ 555 Conjugate | Invitrogen/Life | Cat# C-34776 RRID:AB_2313636 |
| Papain dissociation kit | Worthington Biochemical Corp | Cat# LK003150 RRID: N/A |
| Micro BCA™ Protein Assay Kit | ThermoFisher Scientific | Cat# #23235 RRID: N/A |
| Amersham ECL Prime Western Blotting Detection Reagent | Cytiva | Cat#RPN2232 RRID: N/A |
| Anti mouse IgM MicroBeads | Miltenyi Biotec | Cat# 130–047-302 RRID:AB_244359 |
| Dead cell removal kit | Miltenyi Biotec | Cat# 130–090-101 RRID: N/A |
| Recombinant Wnt3a | R&D Systems | Cat# 5036 RRID: N/A |
| DMSO | Sigma | Cat# D8418 RRID: N/A |
| DMEM | Wisent | Cat# 319–005 RRID: N/A |
| HBSS | Gibco | Cat# 24020–117 RRID: N/A |
| Heat-inactivated horse serum | Gibco | Cat# 16050130 RRID: N/A |
| L-glutamine | Gibco | Cat# 35050–061 RRID: N/A |
| HEPES | Gibco | Cat# 15630–080 RRID: N/A |
| Pen-Strep | Wisent | Cat# 450–201 RRID: N/A |
| PBS | Wisent | Cat# 311–420 RRID: N/A |
| Trypsin | Wisent | Cat# 325–542 RRID: N/A |
| bpV(pic) | Selleckchem | Cat# S8651 |
| BDNF | Peprotech | Cat# 450–02 |
| CNTF | Peprotech | Cat# 450–13 |
| TGF-β1 | Peprotech | Cat# 100–21 |
| TGF-β2 | Peprotech | Cat# 100–35B |
| Forskolin | Sigma | Cat# F6886 |
| B27 | Thermo Fisher Scientific | Cat# 17504044 |
| N2 | Thermo Fisher Scientific | Cat# 17502048 |
| NAC | Sigma | Cat# A8199 |
| Insulin | Invitrogen | Cat# 12585–014 |
| Na Pyruvate | Invitrogen | Cat# 11360–070 |
| L-glutamine | Invitrogen | Cat# 25030–081 |
| T3 | Sigma | Cat# T6397 |
| Neurobasal | Invitrogen | Cat# 21103–049 |
| Paraformaldehyde Granular | Electron Microscopy Sciences | Cat# 19208 |
| Poly-D-Lysine | Corning | Cat# 354210 |
| Laminin | Corning | Cat# 354232 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNAscope® Multiplex Fluorescent Detection Kit v2 | ACD | Cat# 323110 RRID: N/A |
| Chromium Next GEM Single Cell 5′ Reagent Kit v2 | 10X Genomics | Cat# 1000263 |
|
| ||
| Deposited data | ||
|
| ||
| ProteomeXchange Consortium via the PRIDE [1] partner repository (mass spectrometry data) | This study |
Project accession: PXD036221 Project DOI: 10.6019/PXD036221 |
| Uncropped western blot images | This study | https://doi.org/10.17632/jf4pwztxrj |
| scRNA-seq data- P7 wt retinas | This study | GEO: GSE228191 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mus musculus: CD-1 ® IGS mouse | Charles River | Strain Code: 022 RRID:IMSR_CRL:022 |
| Mus musculus: C57BL/6J | The Jackson Laboratory | Cat# 000664 RRID:IMSR_JAX:000664 |
| Ptenfl allele (Backman et al.)37 | The Jackson Laboratory | Strain #: 006440 |
| B6.129S4-Ptentm1Hwu/J | RRID:IMSR_JAX:006440 | |
| Pax6::Cre driver (Marquardt et al.)38 | The Jackson Laboratory | Strain #:024578 |
| Tg(Pax6-GFP/cre)1Rilm/J | RRID:IMSR_JAX:024578. | |
| fro/fro (Alebrahim et al.)58 | Monzur Murshed | (Alebrahim et al.)58 |
| Dscamdel17 (Fuerst et al.)34 | The Jackson Laboratory | Strain #:008000 |
| B6.CBy-Dscamdel17/RwbJ | RRID:IMSR_JAX:008000) | |
| Lef/Tcf-lacZ reporter mice | The Jackson Laboratory | Strain #:004623 |
| Tg(TCF/Lef1-lacZ)34Efu/J | RRID:IMSR_JAX:004623. | |
| Ctnnblfl mice | The Jackson Laboratory | Strain #022775 |
| B6(Cg)-Ctnnb1tm1Knw/J | RRID:IMSR_JAX:022775 | |
|
| ||
| Oligonucleotides | ||
|
| ||
| RNAscope probe: Mm-Pten | ACD | Cat# 316301-C1 |
| RNAscope probe: Mm-Dscam | ACD | Cat# 834031-C2 |
| RNAscope probe: Mm-Th | ACD | Cat# 317621-C3 |
| RNAscope probe: Mm-lsl1 | ACD | Cat# 451931-C3 |
| RNAscope probe: Mm-Smpd3 | ACD | Cat# 815591-C3 |
| Genotyping Pax6::Cre mutant forward: | This study | N/A |
| 5′-CCACGACCAAGTGACAGCAATG-3′ and mutant reverse | ||
| 5′- CAGAGACGGAAATCCATCGCTC-3′ | ||
| Expected band size: MT: 373 bp | ||
| Genotyping primers: Dscam mutant forward | This study | N/A |
| 5′-CTTTGCGCGTTATGATCCT-3′ and Dscam mutant reverse | ||
| 5′-GTGGTGTCGATACTGATG-3′ | ||
| Expected band size: WT: 170 bp MT: 133 bp | ||
| Genotyping primers: Pten flox forward | This study | N/A |
| 5′-CTCCTCTACTCCATTCTTCCC-3′ and Pten flox reverse | ||
| 5′-ACTCCCACCAATGAACAAAC-3′ | ||
| Expected band size: WT: 288 bp MT: 335 bp | ||
|
| ||
| Software and algorithms | ||
|
| ||
| Fiji (ImageJ) | Schindelin et al.78 | https://imagej.net/Fiji |
| GraphPad Prism | GraphPad Software |
https://www.graphpad.com/ RRID:SCR_002798 |
| Seurat v.3.2.3 | Butler et al.79 | https://satijalab.org/seurat |
| MASCOT software version 2.6 (Matrix Science, UK) | Matrix Science | http://www.matrixscience.com/mascot_support_v2_6.html |
| Scaffold version 4.8.2 (USA) | Proteome Software | https://www.proteomesoftware.com/products/scaffold-5 |
| ClueGO Cytoscape plug-in | Bindea et al.80 | http://www.ici.upmc.fr/cluego/cluegoDownload.shtml |
| pathfind R Bioconductor package | Ulgen et al.81 | N/A |
| Qiagen Ingenuity Pathway Analysis (Qiagen IPA) | Qiagen | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ |
| rrvgo Bioconductor package | https://bioconductor.org/packages/release/bioc/vignettes/rrvgo/inst/doc/rrvgo.html | |
| Leica Application Suite X (LASX) | Leica Microsystems | LAS X Life Science Microscope Software - Downloads | Products | Leica Microsystems (leica-microsystems.com) |
| Adobe Photoshop 2021 | Adobe Creative Cloud | Creative business solutions | Adobe Creative Cloud for teams |
| Biorender Science Figures Licence | Biorender | https://biorender.com/ |
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-ca/product/image-lab-software?ID=KRE6P5E8Z |
Highlights.
Pten and Dscam act additively to regulate starburst amacrine cell spacing
Endocytic recycling of cell adhesion molecules is perturbed in PtencKO retinas
Vesicular proteome “fingerprints” endocytic recycling changes in PtencKO retinas
Perturbation of Wnt signaling phenocopies defects in amacrine cell positioning
ACKNOWLEDGMENTS
We thank Rachel O. Wong (University of Washington) for critical discussions, Natasha Klenin (University of Calgary), Petia Stefanova (SRI Histology Facility), Dawn Zinyk (Sunnybrook Research Institute), and Dennis Lee (Sunnybrook Research Institute) for technical support and Sarah Dalesman (University of Calgary) for assistance with statistical analyses of DSCAM+ puncta. We also thank Jung-Lynn (Jonathan) Yang for assistance with artwork (University of Calgary). This work was supported by the Canadian Institutes of Health Research (CIHR) MOP-142338 and CIHR PJT 180243 (to C.S., Y.T., and I.K.) and by the National Institutes of Health (NIH) (EY-019968 to B.E.R.). J.H. was supported by a Canada Graduate Scholarship – Doctoral (CGS-D)/ CIHR award, Vision Science Research Program Scholarship, Ontario Graduate Scholarship, and R.O. Torrance Bursary. N.T. and R.C. were supported by studentships from the ACHRI/CIHR Training Grant at the University of Calgary and by a Lion Sight Center award. C.S. holds the Dixon Family Chair in Ophthalmology Research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests Received: September 8, 2022
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114005.
REFERENCES
- 1.Wässle H. (2004). Parallel processing in the mammalian retina. Nat. Rev. Neurosci. 5, 747–757. [DOI] [PubMed] [Google Scholar]
- 2.Galli-Resta L, Leone P, Bottari D, Ensini M, Rigosi E, and Novelli E. (2008). The genesis of retinal architecture: an emerging role for mechanical interactions? Prog. Retin. Eye Res. 27, 260–283. [DOI] [PubMed] [Google Scholar]
- 3.Galli-Resta L. (2002). Putting neurons in the right places: local interactions in the genesis of retinal architecture. Trends Neurosci. 25, 638–643. [DOI] [PubMed] [Google Scholar]
- 4.Whitney IE, Keeley PW, St John AJ, Kautzman AG, Kay JN, and Reese BE (2014). Sox2 regulates cholinergic amacrine cell positioning and dendritic stratification in the retina. J. Neurosci. 34, 10109–10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deans MR, Krol A, Abraira VE, Copley CO, Tucker AF, and Goodrich LV (2011). Control of neuronal morphology by the atypical cadherin Fat3. Neuron 71, 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galli-Resta L, Resta G, Tan SS, and Reese BE (1997). Mosaics of islet-1-expressing amacrine cells assembled by short-range cellular interactions. J. Neurosci. 17, 7831–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli-Resta L. (2000). Local, possibly contact-mediated signalling restricted to homotypic neurons controls the regular spacing of cells within the cholinergic arrays in the developing rodent retina. Development 127, 1509–1516. [DOI] [PubMed] [Google Scholar]
- 8.Reese BE, and Galli-Resta L. (2002). The role of tangential dispersion in retinal mosaic formation. Prog. Retin. Eye Res. 21, 153–168. [DOI] [PubMed] [Google Scholar]
- 9.Reese BE (2008). Mosaic architecture of the mouse retina. In Eye, Retina and Visual Systems of the Mouse, C. LM., ed. (MIT Press; ), pp. 147–155. [Google Scholar]
- 10.Kozlowski C, Hadyniak SE, and Kay JN (2023). Retinal neurons establish mosaic patterning by excluding homotypic somata from their dendritic territory. Preprint at bioRxiv. 10.1101/2023.11.17.567616. [DOI] [Google Scholar]
- 11.Taylor WR, and Smith RG (2012). The role of starburst amacrine cells in visual signal processing. Vis. Neurosci. 29, 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masland RH (2012). The tasks of amacrine cells. Vis. Neurosci. 29, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockhill RL, Euler T, and Masland RH (2000). Spatial order within but not between types of retinal neurons. Proc. Natl. Acad. Sci. USA 97, 2303–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitney IE, Keeley PW, Raven MA, and Reese BE (2008). Spatial patterning of cholinergic amacrine cells in the mouse retina. J. Comp. Neurol. 508, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reese BE, Necessary BD, Tam PP, Faulkner-Jones B, and Tan SS (1999). Clonal expansion and cell dispersion in the developing mouse retina. Eur. J. Neurosci. 11, 2965–2978. [DOI] [PubMed] [Google Scholar]
- 16.Reese BE, and Tan SS (1998). Clonal boundary analysis in the developing retina using X-inactivation transgenic mosaic mice. Semin. Cell Dev. Biol. 9, 285–292. [DOI] [PubMed] [Google Scholar]
- 17.Cameron DA, and Carney LH (2004). Cellular patterns in the inner retina of adult zebrafish: quantitative analyses and a computational model of their formation. J. Comp. Neurol. 471, 11–25. [DOI] [PubMed] [Google Scholar]
- 18.Tyler MJ, Carney LH, and Cameron DA (2005). Control of cellular pattern formation in the vertebrate inner retina by homotypic regulation of cell-fate decisions. J. Neurosci. 25, 4565–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raven MA, Eglen SJ, Ohab JJ, and Reese BE (2003). Determinants of the exclusion zone in dopaminergic amacrine cell mosaics. J. Comp. Neurol. 461, 123–136. [DOI] [PubMed] [Google Scholar]
- 20.Poché RA, Raven MA, Kwan KM, Furuta Y, Behringer RR, and Reese BE (2008). Somal positioning and dendritic growth of horizontal cells are regulated by interactions with homotypic neighbors. Eur. J. Neurosci. 27, 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenkamp DL, and Cameron DA (2002). Cellular pattern formation in the retina: retinal regeneration as a model system. Mol. Vis. 8, 280–293. [PubMed] [Google Scholar]
- 22.Pollerberg GE, Thelen K, Theiss MO, and Hochlehnert BC (2013). The role of cell adhesion molecules for navigating axons: density matters. Mech. Dev. 130, 359–372. [DOI] [PubMed] [Google Scholar]
- 23.O’Sullivan MJ, and Lindsay AJ (2020). The Endosomal Recycling Pathway-At the Crossroads of the Cell. Int. J. Mol. Sci. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Wang Y, Wong JJL, Lim KL, Liou YC, Wang H, and Yu F. (2014). Endocytic pathways downregulate the L1-type cell adhesion molecule neuroglian to promote dendrite pruning in Drosophila. Dev. Cell 30, 463–478. [DOI] [PubMed] [Google Scholar]
- 25.Meltzer H, and Schuldiner O. (2022). Spatiotemporal Control of Neuronal Remodeling by Cell Adhesion Molecules: Insights From Drosophila. Front. Neurosci. 16, 897706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuerst PG, Bruce F, Rounds RP, Erskine L, and Burgess RW (2012). Cell autonomy of DSCAM function in retinal development. Dev. Biol. 361, 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeley PW, Sliff BJ, Lee SCS, Fuerst PG, Burgess RW, Eglen SJ, and Reese BE (2012). Neuronal clustering and fasciculation phenotype in Dscam- and Bax-deficient mouse retinas. J. Comp. Neurol. 520, 1349–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrett AM, Khalil A, Walton DO, and Burgess RW (2018). DSCAM promotes self-avoidance in the developing mouse retina by masking the functions of cadherin superfamily members. Proc. Natl. Acad. Sci. USA 115, E10216–E10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kay JN, Chu MW, and Sanes JR (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 483, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, and Sanes JR (2012). Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantrup R, Dixit R, Palmesino E, Bonfield S, Shaker T, Tachibana N, Zinyk D, Dalesman S, Yamakawa K, Stell WK, et al. (2012). Cell-type specific roles for PTEN in establishing a functional retinal architecture. PLoS One 7, e32795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MF, and Trotman LC (2020). PTEN: Bridging Endocytosis and Signaling. Cold Spring Harb. Perspect. Med. 10, a036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang C, Manes TD, Liu L, Liu K, Qin L, Li G, Tobiasova Z, Kirkiles-Smith NC, Patel M, Merola J, et al. (2019). ZFYVE21 is a complement-induced Rab5 effector that activates non-canonical NF-κB via phosphoinosotide remodeling of endosomes. Nat. Commun. 10, 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuerst PG, Koizumi A, Masland RH, and Burgess RW (2008). Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature 451, 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keeley PW, and Reese BE (2010). Morphology of dopaminergic amacrine cells in the mouse retina: independence from homotypic interactions. J. Comp. Neurol. 518, 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, and Gan L. (2007). Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J. Neurosci. 27, 12707–12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J, Tsao MS, Shannon P, Bolon B, Ivy GO, and Mak TW (2001). Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29, 396–403. [DOI] [PubMed] [Google Scholar]
- 38.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, and Gruss P. (2001). Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43–55. [DOI] [PubMed] [Google Scholar]
- 39.Amano K, Fujii M, Arata S, Tojima T, Ogawa M, Morita N, Shimohata A, Furuichi T, Itohara S, Kamiguchi H, et al. (2009). DSCAM deficiency causes loss of pre-inspiratory neuron synchroneity and perinatal death. J. Neurosci. 29, 2984–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakagami K, Chen B, Nusinowitz S, Wu H, and Yang XJ (2012). PTEN regulates retinal interneuron morphogenesis and synaptic layer formation. Mol. Cell. Neurosci. 49, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jo HS, Kang KH, Joe CO, and Kim JW (2012). Pten coordinates retinal neurogenesis by regulating Notch signalling. EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tachibana N, Touahri Y, Dixit R, David LA, Adnani L, Cantrup R, Aavani T, Wong RO, Logan C, Kurek KC, and Schuurmans C. (2018). Hamartoma-like lesions in the mouse retina: an animal model of Pten hamartoma tumour syndrome. Dis. Model. Mech. 11, dmm031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tachibana N, Cantrup R, Dixit R, Touahri Y, Kaushik G, Zinyk D, Daftarian N, Biernaskie J, McFarlane S, and Schuurmans C. (2016). Pten Regulates Retinal Amacrine Cell Number by Modulating Akt, Tgfbeta, and Erk Signaling. J. Neurosci. 36, 9454–9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haverkamp S, and Wä ssle H. (2000). Immunocytochemical analysis of the mouse retina. J. Comp. Neurol. 424, 1–23. [PubMed] [Google Scholar]
- 45.Yan W, Laboulaye MA, Tran NM, Whitney IE, Benhar I, and Sanes JR (2020). Mouse Retinal Cell Atlas: Molecular Identification of over Sixty Amacrine Cell Types. J. Neurosci. 40, 5177–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Famiglietti EV Jr., and Kolb H. (1976). Structural basis for ON-and OFF-center responses in retinal ganglion cells. Science 194, 193–195. [DOI] [PubMed] [Google Scholar]
- 47.Raven MA, and Reese BE (2002). Horizontal cell density and mosaic regularity in pigmented and albino mouse retina. J. Comp. Neurol. 454, 168–176. [DOI] [PubMed] [Google Scholar]
- 48.Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, and Burgess RW (2009). DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 64, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawauchi T. (2012). Cell adhesion and its endocytic regulation in cell migration during neural development and cancer metastasis. Int. J. Mol. Sci. 13, 4564–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuerst PG, and Burgess RW (2009). Adhesion molecules in establishing retinal circuitry. Curr. Opin. Neurobiol. 19, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuerst PG, Harris BS, Johnson KR, and Burgess RW (2010). A novel null allele of mouse DSCAM survives to adulthood on an inbred C3H background with reduced phenotypic variability. Genesis 48, 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Andrade GB, Kunzelman L, Merrill MM, and Fuerst PG (2014). Developmentally dynamic colocalization patterns of DSCAM with adhesion and synaptic proteins in the mouse retina. Mol. Vis. 20, 1422–1433. [PMC free article] [PubMed] [Google Scholar]
- 53.Colombo M, Raposo G, and Thé ry C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. [DOI] [PubMed] [Google Scholar]
- 54.Zhou J, Benito-Martin A, Mighty J, Chang L, Ghoroghi S, Wu H, Wong M, Guariglia S, Baranov P, Young M, et al. (2018). Retinal progenitor cells release extracellular vesicles containing developmental transcription factors, microRNA and membrane proteins. Sci. Rep. 8, 2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalargyrou AA, Guilfoyle SE, Smith AJ, Ali RR, and Pearson RA (2022). Extracellular vesicles in the retina - putative roles in physiology and disease. Front. Mol. Neurosci. 15, 1042469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hurwitz SN, Olcese JM, and Meckes DG Jr. (2019). Extraction of Extracellular Vesicles from Whole Tissue. J. Vis. Exp. 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, and Ochiya T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alebrahim S, Khavandgar Z, Marulanda J, and Murshed M. (2014). Inducible transient expression of Smpd3 prevents early lethality in fro/fro mice. Genesis 52, 408–416. [DOI] [PubMed] [Google Scholar]
- 59.Dixson AC, Dawson TR, Di Vizio D, and Weaver AM (2023). Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 24, 454–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hallal S, Tűzesi Á, Grau GE, Buckland ME, and Alexander KL (2022). Understanding the extracellular vesicle surface for clinical molecular biology. J. Extracell. Vesicles 11, e12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, Mohamed O, Dufort D, and Wallace VA (2003). Characterization of Wnt signaling components and activation of the Wnt canonical pathway in the murine retina. Dev. Dynam. 227, 323–334. [DOI] [PubMed] [Google Scholar]
- 62.Huang SMA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620. [DOI] [PubMed] [Google Scholar]
- 63.Fabris F. (2021). Conrad Hal Waddington (1905–1975). In Evolutionary Developmental Biology: A Reference Guide, Rosa L. Nuñ o de laand Müller GB, eds. (Springer International Publishing; ), pp. 299–313. [Google Scholar]
- 64.Bizzarri M, Giuliani A, Minini M, Monti N, and Cucina A. (2020). Constraints Shape Cell Function and Morphology by Canalizing the Developmental Path along the Waddington’s Landscape. Bioessays 42, e1900108. [DOI] [PubMed] [Google Scholar]
- 65.Cullen PJ, and Steinberg F. (2018). To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696. [DOI] [PubMed] [Google Scholar]
- 66.Naguib A, Bencze G, Cho H, Zheng W, Tocilj A, Elkayam E, Faehnle CR, Jaber N, Pratt CP, Chen M, et al. (2015). PTEN functions by recruitment to cytoplasmic vesicles. Mol. Cell 58, 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellenbroek SIJ, Iden S, and Collard JG (2012). Cell polarity proteins and cancer. Semin. Cancer Biol. 22, 208–215. [DOI] [PubMed] [Google Scholar]
- 68.Balakrishnan A, Roy S, Fleming T, Leong HS, and Schuurmans C. (2020). The Emerging Role of Extracellular Vesicles in the Glioma Microenvironment: Biogenesis and Clinical Relevance. Cancers 12, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schramm RD, Li S, Harris BS, Rounds RP, Burgess RW, Ytreberg FM, and Fuerst PG (2012). A novel mouse Dscam mutation inhibits localization and shedding of DSCAM. PLoS One 7, e52652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinyoles M, Del Valle-Pérez B, Curto J, Viñas-Castells R, AlbaCastellón L, García de Herreros A, and Duñach M. (2014). Multivesicular GSK3 sequestration upon Wnt signaling is controlled by p120-catenin/cadherin interaction with LRP5/6. Mol. Cell 53, 444–457. [DOI] [PubMed] [Google Scholar]
- 71.Sarin S, Zuniga-Sanchez E, Kurmangaliyev YZ, Cousins H, Patel M, Hernandez J, Zhang KX, Samuel MA, Morey M, Sanes JR, and Zipursky SL (2018). Role for Wnt Signaling in Retinal Neuropil Development: Analysis via RNA-Seq and In Vivo Somatic CRISPR Mutagenesis. Neuron 98, 109–126.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanaka SS, Kojima Y, Yamaguchi YL, Nishinakamura R, and Tam PPL (2011). Impact of WNT signaling on tissue lineage differentiation in the early mouse embryo. Dev. Growth Differ. 53, 843–856. [DOI] [PubMed] [Google Scholar]
- 73.Rizzoli SO (2014). Synaptic vesicle recycling: steps and principles. EMBO J. 33, 788–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrows CM, McCabe MP, Chen H, Swann JW, and Weston MC (2017). PTEN Loss Increases the Connectivity of Fast Synaptic Motifs and Functional Connectivity in a Developing Hippocampal Network. J. Neurosci. 37, 8595–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, and Eng C. (2005). Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 42, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crino PB (2011). A pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 17, 734–742. [DOI] [PubMed] [Google Scholar]
- 77.Veleva-Rotse BO, and Barnes AP (2014). Brain patterning perturbations following PTEN loss. Front. Mol. Neurosci. 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagè s F, Trajanoski Z, and Galon J. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ulgen E, Ozisik O, and Sezerman OU (2019). pathfindR: An R Package for Comprehensive Identification of Enriched Pathways in Omics Data Through Active Subnetworks. Front. Genet. 10, 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alam S, Zinyk D, Ma L, and Schuurmans C. (2005). Members of the Plag gene family are expressed in complementary and overlapping regions in the developing murine nervous system. Dev. Dynam. 234, 772–782. [DOI] [PubMed] [Google Scholar]
- 83.Touahri Y, Adnani L, Mattar P, Markham K, Klenin N, and Schuurmans C. (2015). Non-isotopic RNA In Situ Hybridization on Embryonic Sections. Curr. Protoc. Neurosci. 70, 1.22.1–25. [DOI] [PubMed] [Google Scholar]
- 84.Thery C, Amigorena S, Raposo G, and Clayton A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 85.Chaturvedi CP, Somasundaram B, Singh K, Carpenedo RL, Stanford WL, Dilworth FJ, and Brand M. (2012). Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/ KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc. Natl. Acad. Sci. USA 109, 18845–18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clark BS, Stein-O’Brien GL, Shiau F, et al. (2019). Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 102, 1111–1126.e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodieck RW (1991). The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis. Neurosci. 6, 95–111. [DOI] [PubMed] [Google Scholar]
- 88.Keeley PW, Eglen SJ, and Reese BE (2020). From random to regular: Variation in the patterning of retinal mosaics. J. Comp. Neurol. 528, 2135–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD: 036221 and https://doi.org/10.6019/PXD036221. Uncropped western blot images were deposited in Mendeley data: 10.17632/jf4pwztxrj.1 (https://doi.org/10.17632/jf4pwztxrj). scRNASeq data were deposited on Gene Expression Omnibus with accession number GEO: GSE228191
This paper does not report original code
All other data reported in this study will be shared by the lead contact upon request.


