Extended Data Fig. 1 |. Latent cells uptake fatty acids secreted by reactive astrocytes.
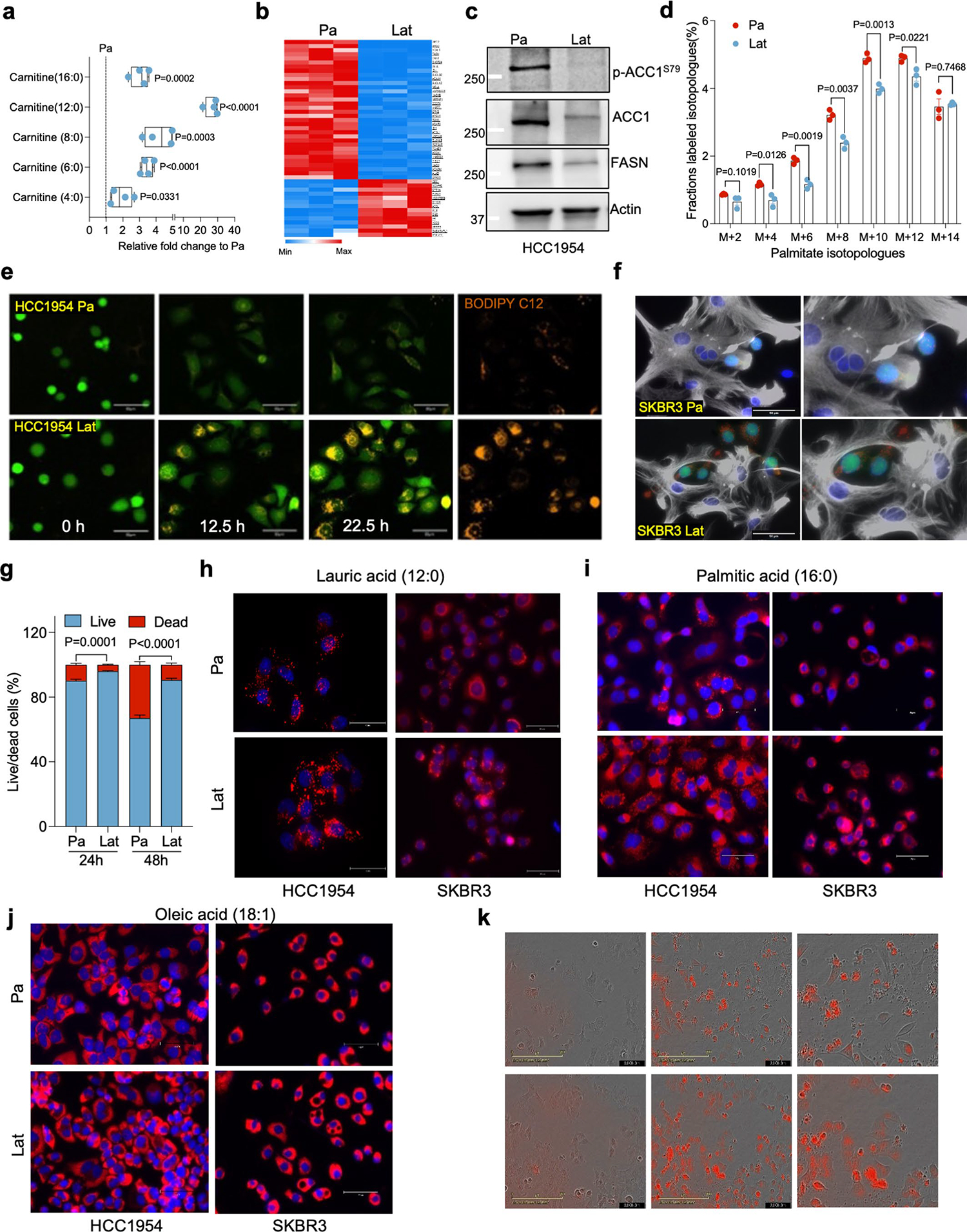
a. Showing relative difference in steady-state carnitine-conjugated fatty acids between Pa and Lat cells. n = 4, each group. Box and whiskers plot showing minima to maxima with all points. b. Heatmap showing differential expression of gene sets related to fatty acid metabolism in HCC1954 Pa and Lat cells. c. Western blots showing expression of fatty acid synthesis related enzymes FASN, ACC1 and p-ACC1S79 in HCC1954 Pa and Lat cells. d. 13C6-glucose tracing data showing the distribution of 13C-labeled even isotopologues of palmitate. n = 3, each group. e. Time-lapse confocal images showing BODIPY-558/568-C12 (Orange) transfer from astrocytes to cancer cells (green) in co-culture setting. f. Immunofluorescence images showing accumulation of LDs (red) in SKBR3 Pa and Lat cells (green) cultured with BODIPY-558/568-C12 labeled reactive astrocytes (Gray, GFAP staining). g. Bar graph showing survival of HCC1954 Pa and Lat cells cultured with astrocytes media. Percentage live/dead cells were quantified at 24 and 48 hours, n = 6, each group. h-j. Nile Red staining IF images showing lipid droplets (red) in HCC1954 and SKBR3 Pa and Lat cells treated with lauric acid, palmitic acid, and oleic acid respectively. Briefly, cells were incubated for 24 hours in R3F media and then treated with 100 μM of indicated FAs for 24 hours. Cells were fixed and stained with Nile red (2 μg/ml) for 10 min. k. IncuCyte3 time-lapse images (0,24 and 48 hours) showing lipid uptake and cell death in HCC1954 cells cultured with 2 μM of BODIPY-558/568-C12. In a, d, and g, ‘n’ represents biologically independent samples and data presented as mean + /−SEM. P value in a, d, and g were calculated by two-tailed Unpaired t-test. The experiments shown in c, e, f, and h-k were repeated independently at least two or three times with similar results.
