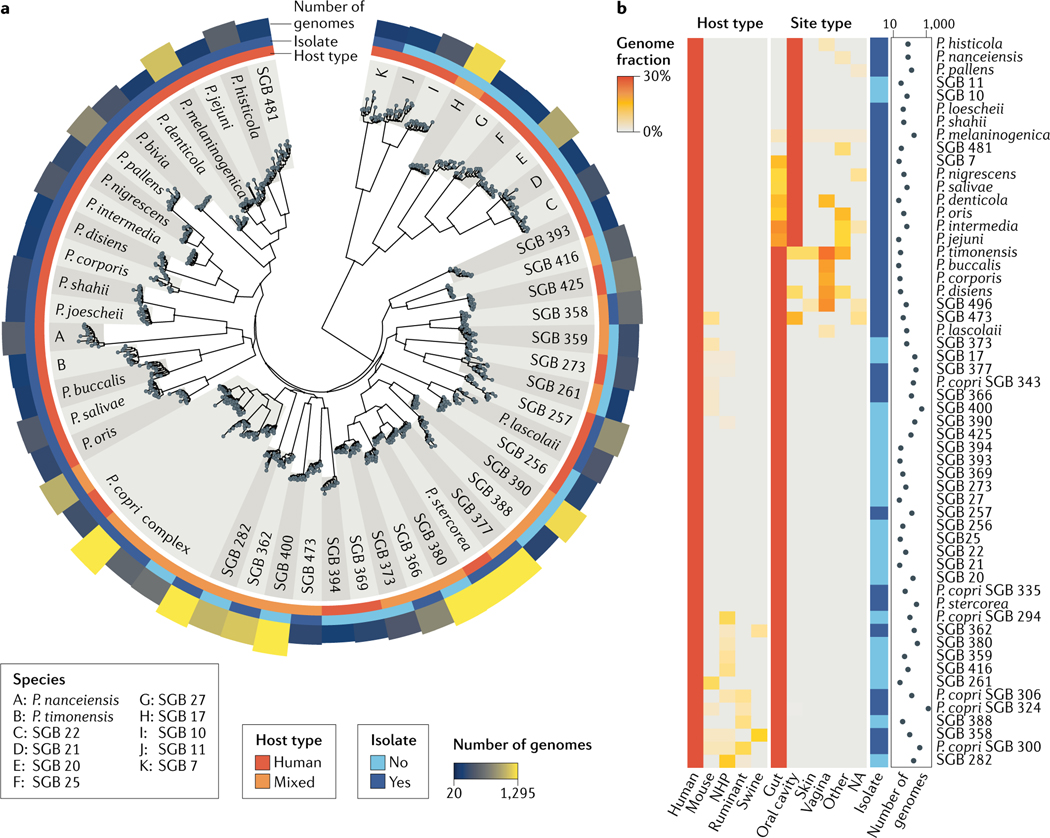
Fig. 3 |. Current diversity of Prevotella spp. in the human microbiome.
a | A large phylogenetic tree spanning the 56 most prevalent Prevotella species (with at least 20 genomes retrieved from human microbiomes). Isolate sequences were integrated with metagenome-assembled genomes (MAGs) retrieved from more than 10,000 metagenomes and more than 50 human populations. For each species, 15 randomly selected genomes are reported along with the information on the host type (that is, human or mixed sources), the public availability of isolate sequences and the number of retrieved genomes. The tree highlights the gap in strain isolation, with 24 human-associated species that completely lack isolate genomes, the within-species strain diversity and the remarkably well-defined taxonomic species based on the interspecies versus the intraspecies diversity. b | The fraction of Prevotella genomes retrieved from human and non-human hosts (both from isolation and assignment to MAGs) and different human site types suggest multiple ecological patterns. In addition to human-specific species, other species are present in two or more host types. Of note, a few species are ecologically adapted to multiple human body sites, which might also be due to the influx of oral species into the lower gastrointestinal tract71. Supplementary Table 1 provides a list of available reference genomes and reconstructed MAGs with their genomic characteristics for the species considered in this figure. NA, not assigned; NHP, non-human primate; SGB, species-level genome bin.