Abstract
Hepatitis E virus (HEV) is an unclassified virus with a positive-sense RNA genome and an undefined replication strategy. In order to determine whether the HEV genome is capped or not, we developed a reverse transcription-PCR assay that is based on the ability of a monoclonal antibody to recognize 7-methylguanosine (m7G). Antibody to m7G bound RNA extracted from virions of two different HEV genotypes. The cap analog competitively inhibited the binding of virion RNAs, demonstrating that HEV has a capped RNA genome.
Hepatitis E virus (HEV) is a spherical, nonenveloped virus, 30 to 32 nm in diameter. HEV was initially classified as a member of the family Caliciviridae (2) but was recently removed from this family and is again unclassified (8). The positive-sense single-stranded RNA genome of approximately 7.5 kb is organized into three open reading frames with a short 5′ nontranslated region (NTR) and a short 3′ NTR terminated by a poly(A) tract (9, 11). The largest open reading frame, ORF1, begins 27 nucleotides (nt) downstream of the apparent 5′ end of the genome and extends about 5 kb. It is presumed to encode nonstructural proteins, since it has motifs characteristic of several viral enzymes, including a methyltransferase, a papain-like cysteine protease, an RNA helicase, and an RNA-dependent RNA polymerase. ORF2, encoding the putative capsid protein, begins downstream of ORF1 and extends approximately 2,000 nt before terminating upstream of the polyadenylation site. ORF3, less than 400 nt in length, overlaps both ORF1 and ORF2 and encodes a small immunogenic phosphoprotein which associates with the cytoskeleton (6, 14, 17).
The replication strategy of HEV is still not understood (12). The presence of a methyltransferase motif in ORF1 suggests that HEV may have a capped RNA genome, since this enzyme is generally responsible for methylating the 5′-terminal guanosine in the cap to produce the structure m7G(5′)ppp(5′)X (13). HEV has not been available in quantities sufficient for biochemical characterization, and the lack of an efficient cell culture system prevents radioactive labeling experiments that might provide direct evidence for a cap at the 5′ end of the genome.
Antibody to cap structure has been used previously in immunoadsorption assays to demonstrate that RNAs of some viruses are capped (4, 5, 10). Therefore, in order to determine whether the HEV genome is capped or not, we have developed a reverse transcription-PCR (RT-PCR) assay that is based on the ability of a monoclonal antibody (MAb) to 2,2,7-trimethyl guanosine (m3G) to cross-react with intact m7G cap structures (1). This antibody was previously shown to precipitate m7G-capped SP6–β-globin RNA but not GpppG-terminated or noncapped RNAs (1).
Synthesis of control RNAs.
The full-length genome of HEV, strain SAR-55, was amplified by long RT-PCR essentially as described by Tellier et al. (15). Briefly, genomic RNA was extracted from HEV in 200 μl of 10% human stool suspension with Trizol reagent (Gibco-BRL) and reverse transcribed with Superscript II reverse transcriptase (Gibco-BRL) at 42°C for 1 h. The cDNA was subsequently amplified by using the KlenTaq advantage kit (Clontech); the forward primer 5′-GGGTC TAGA(TAATACGACTCACTATA)GAGGCAGACCACAT ATG-3′, comprising an XbaI site (boldface and underlined), a T7 promoter (parentheses), and the terminus of HEV 5′ noncoding region; and the reverse primer 5′-GGGGATATCTTTTTTTTTTTTTTTCAGGGAGCGCGGAACG-3′, comprising an EcoRV site (boldface and underlined), a poly(T) tract, and the terminus of HEV 3′ noncoding region. The gel-purified HEV cDNA and the vector PSK(+) II (Stratagene) were both digested with XbaI and EcoRV and ligated together to provide a full-length clone. The full-length clone of HEV was digested with EcoNI and SnaBI, to delete a fragment of about 100 bp in the 5′ half of ORF1 (positions 1508 to 1608). Digested DNA was blunt ended with the Klenow fragment of DNA polymerase I (Gibco-BRL) and ligated by T4 DNA ligase (Gibco-BRL) to provide a deleted plasmid. The plasmids containing full-length or deleted HEV genome were purified with the Qiagen Plasmid Maxi kit, linearized with EcoRV, and transcribed in vitro with the AmpliScribe T7 transcription kit (Ambion) in order to generate capped (Epicentre reagents) or uncapped RNAs for controls. After transcription, DNA template was eliminated by digestion with RNase-free DNase I (Promega) for 30 min at 37°C. The transcribed RNAs were purified by LiCl precipitation (Ambion) followed by extraction with Trizol reagent (Gibco-BRL). Dilutions of purified transcribed RNAs were directly subjected to PCR without prior reverse transcription to verify the removal of the DNA template.
Immunoselection of RNAs with anticap MAb.
MAb H20, which recognizes the m3G and m7G cap structures (1), was kindly provided by Reinhard Luhrmann, University of Marburg. Sepharose beads (Gammabind G-Sepharose; Pharmacia) (500 μl) were washed with binding buffer (10 mM NaH2PO4, 150 mM NaCl, 10 mM EDTA, pH 7.0) and incubated with the H20 MAb at 4°C overnight. The antibody-coupled beads were washed three times with binding buffer. As a control for the specificity of RNA binding, beads were also coupled to an irrelevant MAb, antikeratin (Cappel-ICN Pharmaceuticals, Inc.), under the same conditions.
For each reaction, 75 μl of a 50% suspension of beads coupled to anticap or to antikeratin MAb was mixed with 25 μl of in vitro-transcribed RNAs or with authentic HEV virion RNA, diluted to a final volume of 150 μl with binding buffer, and incubated at 4°C for 1 h. After centrifugation, the supernatant was collected and the beads were washed three times in 500 μl of binding buffer containing 0.5% Nonidet P-40. The competition assay was performed by incubating the anticap MAb-coupled beads with the methylated cap analog, m7G(5′)ppp(5′)G, or with unmethylated cap analog, G(5′)ppp(5′)G (Epicentre), for 1 h at 37°C prior to incubation of the washed beads with RNAs. RNA bound to the anticap MAb-coupled beads and free RNA in the supernatant were recovered by phenol extraction and ethanol precipitation. The RNA pellets were suspended in 25 μl of water containing 40 U of RNasin and 0.1% dithiothreitol.
RT-PCR.
Primers specific for the same 5′ portion of ORF1 of the SAR-55 strain (16) and of the swine HEV strain (7) were synthesized commercially (Gibco-BRL). The SAR-55 forward primer (positions 1362 to 1382) and the reverse primer (positions 1705 to 1685) amplified a region spanning the deletion in the transcribed SAR-55 RNA genome. RNAs extracted from anticap MAb-coupled beads and from corresponding supernatants were incubated for 10 min at 65°C and then chilled on ice. Reverse transcription with antisense primer was performed with Superscript II reverse transcriptase (Gibco-BRL) at 42°C for 1 h, and cDNA was amplified by PCR with AmpliTaq Gold polymerase (Perkin-Elmer). The PCR consisted of 39 cycles of denaturation at 94°C for 1 min, annealing at 48°C for 1 min, and extension at 72°C for 1 min 30 s. PCR products were visualized by UV light after electrophoresis in a 2% agarose gel containing ethidium bromide. The band expected from amplification of the in vitro transcript of the full-length HEV clone and from HEV virion RNAs was 343 bp, and that from the in vitro transcript of the deleted clone was 243 bp.
Specificity controls.
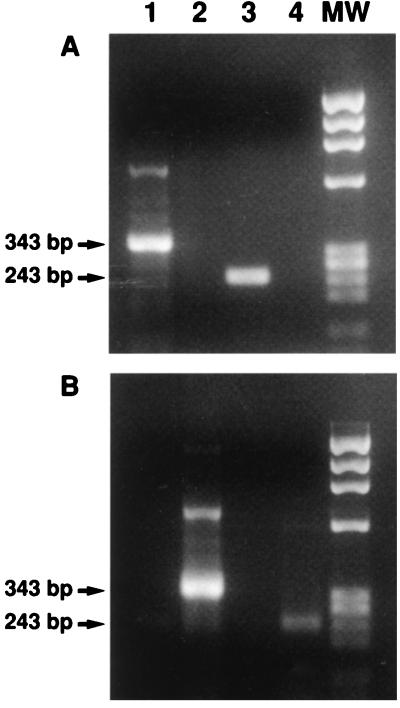
Capped and uncapped SAR-55 RNAs transcribed in vitro were used to determine the optimal conditions for specific binding of viral RNA to anticap MAb coupled to the beads. Semiquantitative RT-PCR of capped and uncapped RNA transcripts was performed to determine the PCR titer, expressed in genome equivalents (GE) for each transcript. One hundred GE of each transcribed RNA was incubated with anticap MAb-coupled beads. A constant volume of beads coupled to anticap or antikeratin MAb was incubated with an equivalent amount of each capped or uncapped transcript. After incubation and centrifugation, RNA was extracted from the MAb-coupled beads and from the corresponding supernatant, and the paired RNA samples were subjected to RT-PCR at the same time under the same conditions. Neither capped nor uncapped transcripts bound to the beads coupled to the antikeratin (data not shown). The RNA extracts from the anticap MAb-coupled beads were positive for HEV genome after incubation with capped RNA transcripts, whereas the corresponding supernatants were negative. In contrast, the same beads incubated with uncapped RNA transcripts were negative for HEV genomes, and the corresponding supernatants were positive (Fig. 1).
FIG. 1.
Capped RNA controls are selectively bound to anticap MAb coupled to Sepharose beads. Shown are RT-PCR products of HEV RNAs extracted from anticap-coupled beads (A) or supernatants (B) following incubation with capped in vitro transcripts (lanes 1 and 3) or uncapped in vitro transcripts (lanes 2 and 4). MW, molecular weight markers; 343 bp, full-length-genome PCR product; 243 bp, deleted-genome PCR product.
Binding of HEV genomic RNA by anticap antibody.
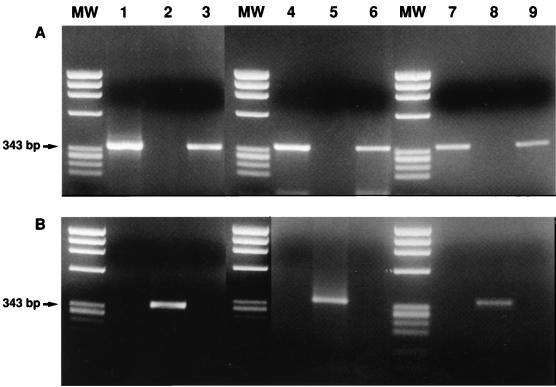
HEV genomic RNA was extracted from a human stool suspension containing the SAR-55 strain (16) and from a rhesus monkey stool suspension containing the swine HEV strain (7). One hundred GE was similarly incubated with anticap MAb-coupled beads. The beads and the corresponding supernatant were extracted as described for the in vitro RNA transcripts. As shown in Fig. 2, HEV cDNA from the human HEV strain as well as from the swine HEV strain was amplified from the extract of the beads but not from the extract of the corresponding supernatant, demonstrating that HEV virion RNA was bound by the anticap MAb. This experiment was repeated many times with similar results. The specificity of the binding reaction was also confirmed by modifying the experiment to include an additional step in which the anticap MAb-coupled beads were incubated with an excess amount of methylated cap analog or unmethylated cap analog before the addition of HEV RNAs. The unmethylated cap analog did not compete for binding (Fig. 2). In contrast, the binding of authentic virion RNAs and capped in vitro-transcribed control RNA was abolished by prior incubation of the MAb-coupled beads with methylated cap analog (Fig. 2).
FIG. 2.
Cap analog blocks the binding of capped control RNA and HEV genomic RNA by anticap MAb. Shown are RT-PCR products of HEV RNAs extracted from anticap-coupled beads (A) or supernatants (B) following incubation with capped RNA transcript (lanes 1 to 3), virion RNA extracted from the human HEV strain (lanes 4 to 6), or virion RNA from the swine HEV strain (lanes 7 to 9); the beads were preincubated with m7(5′)GpppG (lanes 2, 5, and 8) or GpppG (lanes 3, 6, and 9). MW, molecular weight markers.
These data strongly support the conclusion that the HEV genome is indeed capped. The H20 MAb used in this experiment is known to have a high affinity for both m3G and m7G cap structures (1). Synthetic HEV genomic RNA bound the H20 MAb-coupled beads only when it was capped. The two HEV strains tested are as distant as any two HEV strains and have only 79 to 80% nucleotide sequence homology in the ORF2 gene (7), but they reacted in the assay in a manner identically to that of each other and of the in vitro-capped RNA. Moreover, binding could be competitively inhibited by the cap analog m7G(5′)ppp(5′)G, confirming that binding was specific for the methylated cap structure. The conclusion that the HEV genome is capped is also supported by sequence data that assign a very short (27 nt) 5′ NTR to the viral genome. A short NTR is compatible with translation directed by a cap but not by an internal ribosome entry site. Although the completeness of the 5′ terminus of HEV cannot be confirmed until the infectivity of a cDNA is demonstrated, attempts to detect additional 5′ nucleotides have failed, suggesting that the entire sequence has been identified (3, 6a). Therefore, attempts to construct an infectious cDNA clone of HEV should include provisions for a cap.
Acknowledgments
We are most grateful to R. Luhrmann for providing the H20 cap MAb. We thank Judith Graff for helpful discussions.
REFERENCES
- 1.Bochnig P, Reuter R, Bringmann P, Luhrmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact U sn RNPs as well as with 7-methylguanosine. Eur J Biochem. 1987;168:461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- 2.Cubitt D, Bradley D W, Carter M J, Chiba S, Estes M K, Saif L J, Schaffer F L, Smith A W, Studdert M J, Thiel H J. Caliciviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Archives of virology, supplement 10. Vienna, Austria: Springer-Verlag; 1995. pp. 359–363. [Google Scholar]
- 3.Donati M C, Fagan E A, Harrison T J. Sequence analysis of full-length HEV clones derived directly from human liver in fulminant hepatitis E. In: Rizzetto M, Purcell R H, Gerin J L, Verme G, editors. Viral hepatitis and liver diseases. Turin, Italy: Edizioni Minerva Medica; 1997. pp. 313–316. [Google Scholar]
- 4.Garcin D, Kolakofsky D. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J Virol. 1990;64:6196–6203. doi: 10.1128/jvi.64.12.6196-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacker D, Rochat S, Kolakofsky D. Anti-mRNAs in La Crosse bunyavirus-infected cells. J Virol. 1990;64:5051–5057. doi: 10.1128/jvi.64.10.5051-5057.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jameel S, Zafrullah M, Ozdener M H, Panda S K. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol. 1996;70:207–216. doi: 10.1128/jvi.70.1.207-216.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Meng, X. J. Unpublished data.
- 7.Meng X J, Purcell R H, Halbur P G, Lehmans J R, Webb D M, Tsareva T S, Haynes J S, Thacker B J, Emerson S U. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pringle C R. Virus taxonomy—San Diego 1998. Arch Virol. 1998;143:1449–1459. doi: 10.1007/s007050050389. [DOI] [PubMed] [Google Scholar]
- 9.Purcell R H. Hepatitis E virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2831–2843. [Google Scholar]
- 10.Ramirez B C, Garcin D, Calvert L A, Kolakofsky D, Haenni A L. Capped nonviral sequence at the 5′ end of the mRNAs of rice hoja blanca virus RNA4. J Virol. 1995;69:1951–1954. doi: 10.1128/jvi.69.3.1951-1954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes G R, Purdy M A, Kim J P, Luk K C, Young L M, Fry K E, Bradley D W. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1336–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 12.Reyes G R, Huang C C, Tam A W, Purdy M A. Molecular organization and replication of hepatitis E virus (HEV) Arch Virol. 1993;7(Suppl.):15–25. doi: 10.1007/978-3-7091-9300-6_2. [DOI] [PubMed] [Google Scholar]
- 13.Shatkin A J. Capping of eukaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 14.Tam A W, Smith M M, Guerra M E, Huang C, Bradley D W, Fry K E, Reyes G R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–130. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tellier R, Bukh J, Emerson S U, Purcell R H. Amplification of the full-length hepatitis A virus genome by long reverse transcription-PCR and transcription of infectious RNA directly from the amplicon. Proc Natl Acad Sci USA. 1996;93:4370–4373. doi: 10.1073/pnas.93.9.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsarev S A, Emerson S U, Reyes G R, Tsareva T S, Legters L J, Malik I A, Iqbal M, Purcell R H. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zafrullah M, Ozdener M H, Panda S K, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol. 1997;71:9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]