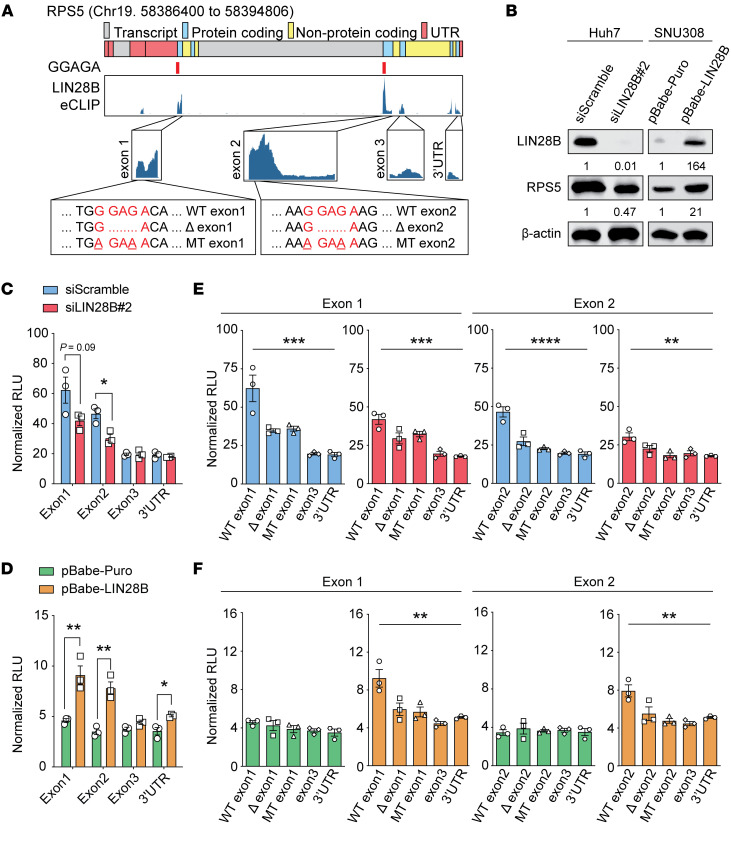
Figure 5. LIN28B regulates RPS5 translation through direct mRNA binding.
(A) eCLIP data show LIN28B-binding regions on RPS5 mRNA. Red marks under the gene indicate the location of consensus LIN28B-binding motifs (GGAGA). Deletion (Δ) and mutation (MT) of consensus motifs were introduced to prevent LIN28B binding. Exon 1, 2, 3, and 3′ UTR sequences were inserted into the Renilla 3′ UTR region. (B) WB analysis of RPS5 and LIN28B in Huh7 and SNU308 cell lines. Numbers below the boxes show relative intensities. (C) Renilla luciferase activity promoted by RPS5 exon 1, exon 2, exon 3, and 3′ UTR reporters in LIN28B siRNA knockdown (n = 3) versus control Huh7 cells (n = 3). (D) Renilla luciferase activity promoted by RPS5 exon 1, exon 2, exon 3, and 3′ UTR reporters in LIN28B overexpression (n = 3) versus control SNU308 cells (n = 3). (E and F) Renilla luciferase activity promoted by WT RPS5 sequences compared with deletion and mutation containing reporters in control (blue) and LIN28B siRNA knockdown (red) Huh7 cells (n = 3) (E) and in control overexpression (green) and LIN28B overexpression (orange) SNU308 cells (n = 3) (F). One-way ANOVA was performed. **P < 0.01; ***P < 0.001; ****P < 0.0001.