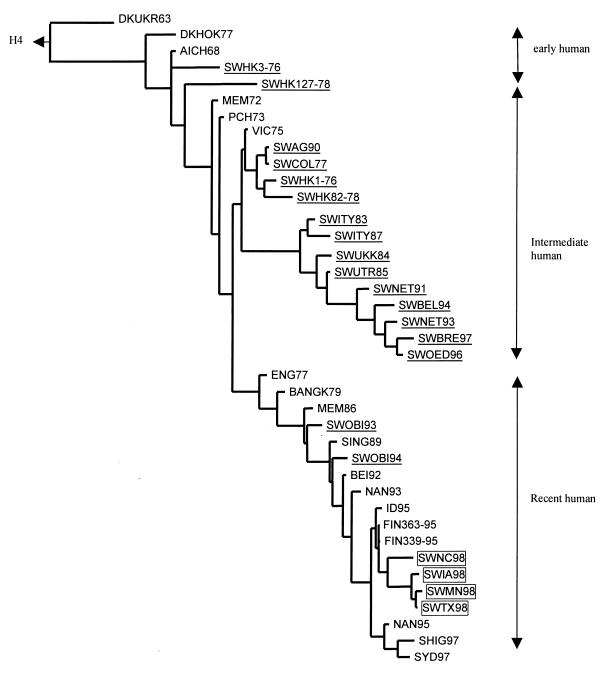
FIG. 1.
Phylogenetic tree of HA1 nucleotide sequences of the H3 hemagglutinin. The tree was generated by the maximum parsimony method in the PAUPsearch and PAUPdisplay programs of the Wisconsin Package version 9.1-Unix (Genetics Computer Group) and is rooted to the H4 sequence of A/Duck/Czechoslovakia/56 (H4N6). Horizontal lines are proportional to the numbers of nucleotide substitutions between branch points. Swine viruses are underlined, and the SW98 viruses are boxed. Abbreviations: DKUKR63, A/Duck/Ukraine/1/63 (H3N8); DKHOK77, A/Duck/Hokkaido/5/77 (H3N?); AICH68, A/Aichi/2/68 (H3N2); MEM72, A/Memphis/102/72 (H3N2); PCH73, A/Port Chalmers/1/73 (H3N2); VIC75, A/Victoria/3/75 (H3N2); ENG77, A/England/321/77 (H3N2); BANGK79, A/Bangkok/1/79 (H3N2); MEM86, A/Memphis/6/86 (H3N2); SING89, A/Singapore/12/89 (H3N2); BEI92, A/Beijing/32/92 (H3N2); NAN93, A/Nanchang/12/93 (H3N2); NAN95, A/Nanchang/933/95 (H3N2); FIN363-95, A/Finland/363/95 (H3N2); FIN339-95, A/Finland/339/95 (H3N2); ID95, A/Idaho/4/95 (H3N2); SHIG97, A/Shiga/25/97 (H3N2); SYD97, A/Sydney/05/97 (H3N2); SWHK1-76, A/Swine/Hong Kong/1/76 (H3N2); SWHK3-76, A/Swine/Hong Kong/3/76 (H3N2); SWCOL77, A/Swine/Colorado/1/77 (H3N2); SWHK82-78, A/Swine/Hong Kong/82/78 (H3N2); SWHK127-78, A/Swine/Hong Kong/127/78 (H3N2); SWITY83, A/Swine/Italy/309/83 (H3N2); SWUKK84, A/Swine/Ukkel/1/84 (H3N2); SWUTR85, A/Swine/Utrecht/4/85 (H3N2); SWITY87, A/Swine/Italy/635/87 (H3N2); SWAG90, A/Swine/Ange-Gardien/150/90 (H3N2); SWNET91, A/Swine/Netherlands/784/91 (H3N2); SWNET93, A/Swine/Netherlands/L2/93 (H3N2); SWOBI93, A/Swine/Obihiro/1/93 (H3N2); SWOBI94, A/Swine/Obihiro/1/94 (H3N2); SWBEL94, A/Swine/Belgium/231/94 (H3N2); SWOED96, A/Swine/Oedenrode/7C/96 (H3N2); SWBRE97, A/Swine/Breugel/97 (H3N2).