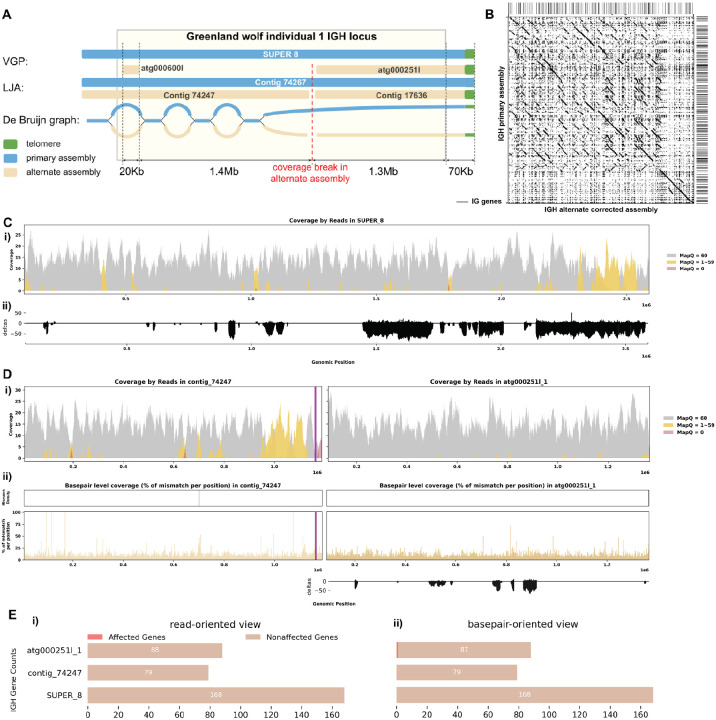
Figure 4. Reassembly Analysis of IGH Locus Assembly Errors in C. lupus individual 1.
A. Primary and alternate assemblies of C. lupus IGH locus produced by VGP and LJA are represented by blue (primary) and tan (alternate) rectangles. Telomere sequences (tandem repeats with repeating sequence CCCTAA) are shown in green. B. Dotplots comparing gene locations and alignments are shown for the corrected alternate IGH assembly vs primary IGH assembly. C. Re-analysis of alignment mismatch in the primary IGH haplotype, i) read coverage across the entire IGH loci, color-coded by mapping quality, and ii) basepair-oriented mismatch rate, a heatmap above indicating the frequency of high mismatch rate base pairs, with darker colors denoting more frequent occurrences. Light red highlights positions covered by ≥5 reads with an error rate >1%, and purple bars indicate coverage breaks (coverage ≤2). D. Similar analyses are performed for the corrected alternate haplotype. E. IG gene quality from i) read-oriented view, affected genes are IGH genes covered by > 5 poorly aligned reads; and ii) basepair-oriented view, affected genes are IGH genes without perfect support (>80% confidence at every position)