Fig. 3. Mapping high-resolution eQTLs.
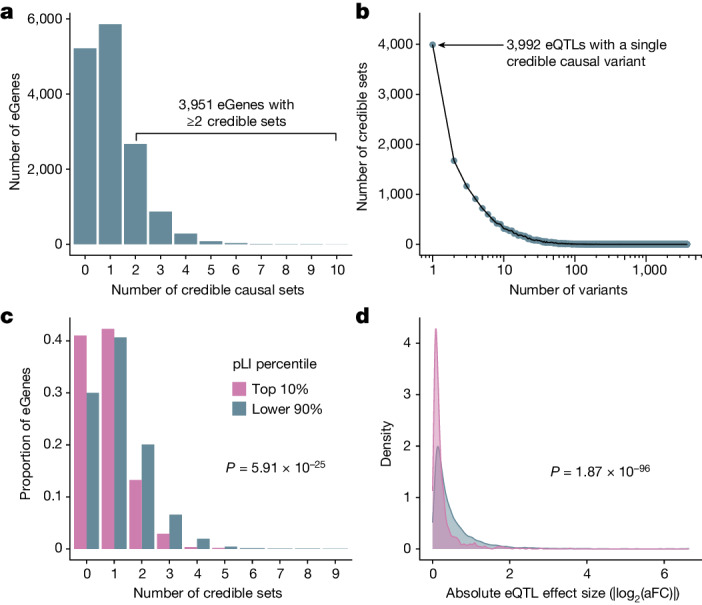
a, Number of credible sets per eGene, demonstrating evidence of widespread allelic heterogeneity, whereby multiple causal variants independently modulate expression of the same genes. b, Fine-mapping resolution, defined as the number of variants per credible set. c, A signature of negative selection against expression-altering variation, whereby eGenes under strong evolutionary constraint (defined as the top pLI decile reflecting intolerance to loss-of-function mutations; pink) possess fewer credible sets, on average, than other genes (blue). d, A signature of negative selection against expression-altering variation, whereby eQTLs of genes under strong evolutionary constraint (top pLI decile; pink) have smaller average effect sizes (aFC) than other genes (blue).
