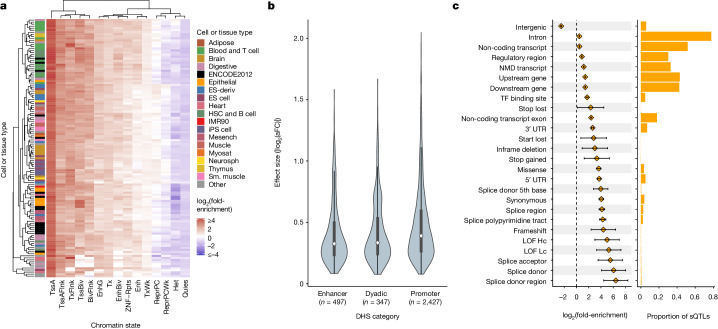
Fig. 4. Fine-mapped cis-QTLs are strongly enriched in regulatory regions across multiple cell and tissue types.
a, A heatmap representing hierarchical clustering of the enrichment of cis-eQTLs in predicted chromatin states using the Roadmap Epigenomics 15-state chromHMM model across 127 cell and tissue samples. b, Distribution of absolute value of lead cis-eQTL effect sizes measured as log2(aFC) across putatively active chromatin states of LCLs linked to multi-tissue DHSs. Sample sizes describe the number of unique eVariants annotated as belonging to each of the DHS categories. Bars represent the first, second (median) and third quartiles of the data and whiskers are bound to 1.5× the interquartile range. c, Enrichment of lead sQTLs (n = 13,107 unique sVariants total, at least 5 per category) within functional annotation categories from Ensembl Variant Effect Predictor (left), along with the proportion of all lead sQTLs falling into each annotation category (right). Error bars denote 95% CI around the estimated sQTL enrichment in each category. Enrichment was calculated in comparison to a background set of variants matched on MAF and distance from the TSS. Annotation categories are not mutually exclusive and therefore sum to a proportion greater than 1. ES, embryonic stem; HSC, haematopoietic stem cell; iPS, induced pluripotent stem; Mesench, mesenchymal cell; Myosat, myosatellite cell; Neurosph, neurosphere; Sm., smooth; TssA, active TSS; TssAFlnk, flanking active TSS; TxFlnk, transcription at gene 5′ and 3′; Tx, strong transcription; TxWk, weak transcription; EnhG, genic enhancer; Enh, enhancer; ZNF/Rpts, ZNF genes plus repeats; Het, heterochromatin; TssBiv, bivalent/poised TSS; BivFlnk, flanking bivalent TSS/enhancer; EnhBiv, bivalent enhancer; ReprPC, repressed polycomb; ReprPCWk, weak repressed polycomb; Quies, quiescent/low; NMD, nonsense-mediated mRNA decay; LOF, loss of function; Hc, high confidence; Lc, low confidence.