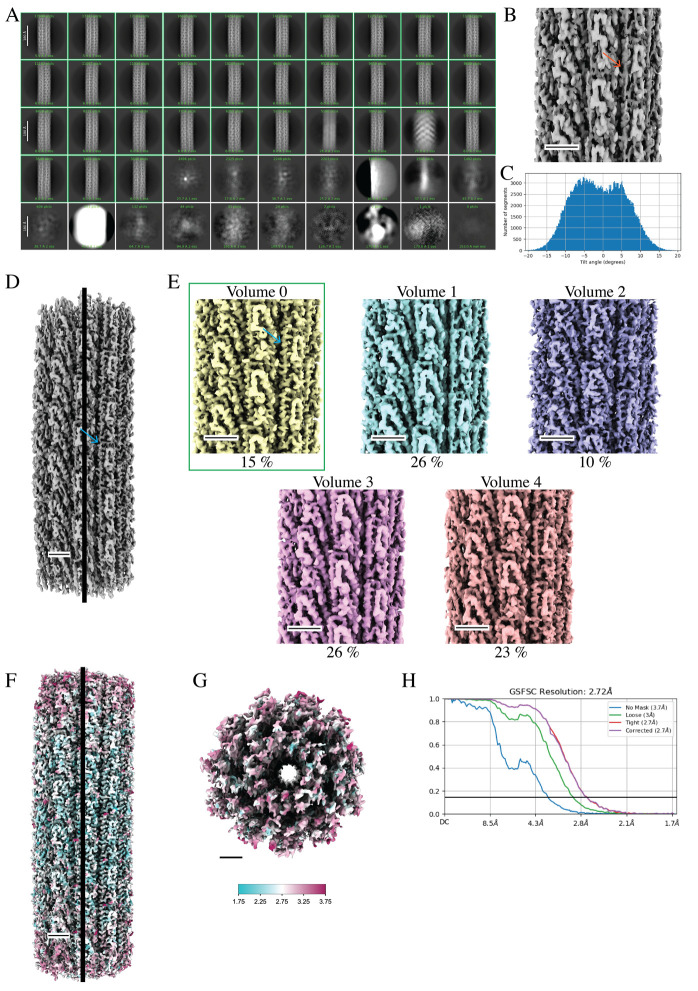
Figure 9. Cryo-EM data processing of curved helical polymers by asymmetrical reconstruction methods improves map quality.
A. 2D classes from particles that were picked after training the filament tracer neural net (step G11). Green boxes highlight classes that were selected for further processing (step G12) (scale bar, 180 Å). B. Initial 3D volume with imposed helical symmetry results in the blurring of secondary structures. The red arrow indicates an expected α-helix that is blurred and more “tube-like” (step G13). C. Bimodal distribution of particle tilt angles suggests that the high curvature observed in the micrographs impedes high-resolution structure determination when imposing helical symmetry (step G13). D. Curved 3D volumes are observed after 3DVA. The best volume is used as a reference for one round of homogeneous refinement (no helical symmetry imposed) using all particles. This process results in a map with much higher quality and improved secondary structure features (blue arrow). Black line indicates the center of the 3D volume. E. A second round of 3DVA helps to sort heterogeneity within the particle set. Volume 0 has the best-resolved α-helices (blue arrow) and is selected (green box) for further processing. F. The selected particle set is used for additional rounds of homogeneous reconstruction, CTF refinement, and local motion correction, resulting in a final map with improved map quality displaying secondary structures and side-chain densities. The local resolution map shows that the highest resolved density is along the center of the helical axis. G. A top view of the filament shows lower resolution density at the end of the reconstruction. The key indicates a map range from 1.75 to 3.75 Å (step G26). H. The GS-FSC reports a global resolution of 2.72 Å. Scale bars for the 3D volumes are 25 Å.