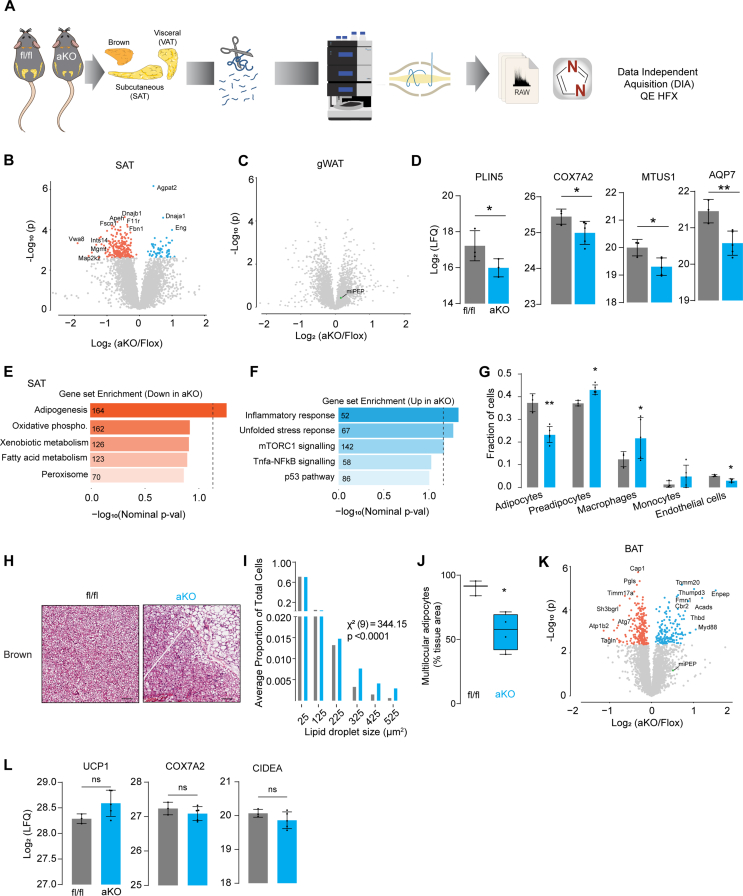
Figure 5.
adipo-miPEP-KO mice do not exhibit adipose tissue browning. (A) Experimental workflow for global proteomics of gonadal white adipose tissue (gWAT), subcutaneous white adipose tissue (SAT) and brown adipose tissue (BAT) in aKO mice. (B–C) Volcano plot of relative protein abundance across adipose tissues. Orange, significatively downregulated proteins. Light blue, significatively upregulated proteins (−log10(p-val) = 2.2). (D) Abundance of proteins associated with browning, N = 3–5, Mean ± S.D. ∗p < 0.05 vs miPEPfl/fl. Gene set enrichment analysis of downregulated (E) and upregulated (F) proteins in adipo-miPEP-KO mice. Number of proteins associated with each pathway is shown within each bar. (G) Cell type deconvolution using proteomic data from SAT whole tissue aKO and miPEPfl/fl (fl/fl) mice. Mean ± S.D., N = 3–5, ∗p < 0.05, ∗∗p < 0.01 vs miPEPfl/fl mice. (H) Haematoxylin and eosin staining of brown adipose tissue (BAT). Scale bar 50 μm. (I) Quantification of the adipocyte size distribution for BAT (N = 5). (J) Percentage of cells with multilocular lipids droplets relative to the whole tissue area. N = 4, Median ± S.D. ∗p < 0.05 vs miPEPfl/fl. (K) Volcano plot of relative protein abundance in BAT. Orange, significatively downregulated proteins. Light blue, significatively upregulated proteins. (L) Abundance of proteins associated with BAT activity, N = 3–5, Mean ± S.D. ∗p < 0.05 vs miPEPfl/fl.