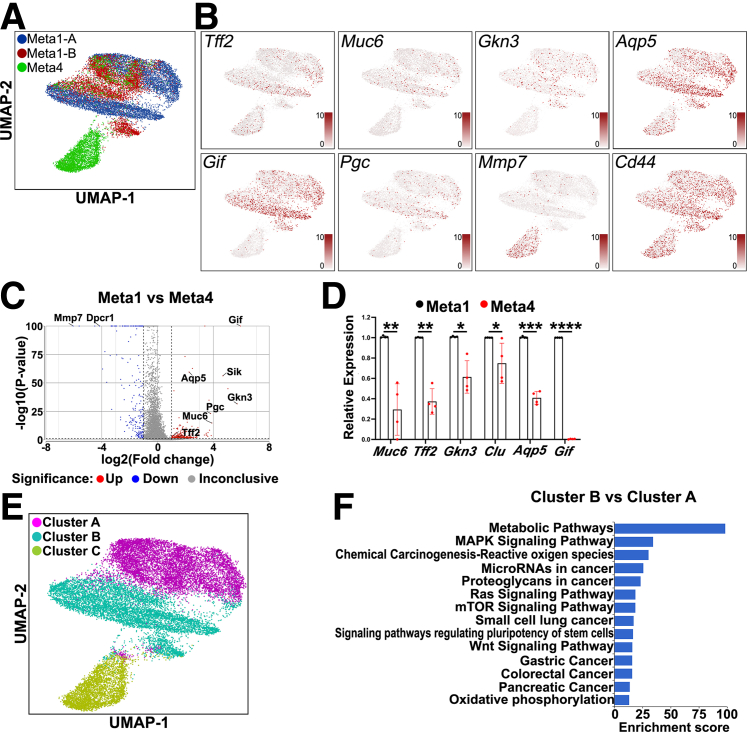
Figure 3.
Single-cell RNA-seq of Meta1 gastroid lines. (A) UMAP plot three-colored to show the cell distribution of the Meta1 and Meta4 gastroid lines. Each dot represents a single cell. (B) UMAP plots showing expression of different genes in red. Tff2, Muc6, Gkn3, and Aqp5 are SPEM-related genes and overlayed with Meta1 gastroid-derived cells. Gif and Pgc are genes expressed in chief cells and overlayed with Meta1 gastroid-derived cells. Mmp7, a dysplastic marker, overlayed with the Meta4 gastroid-derived cells and Cd44 was present in both types of gastroids. (C) Volcano plot of the differentially expressed genes between both Meta1 lines versus Meta4 gastroid line with emphasis in up-regulation of SPEM-related genes and down-regulation of dysplastic-related genes in Meta1 gastroid cells. (D) Quantitative RT-PCR showing the relative expression levels of key chief cell and SPEM cell genes. Mean ± SD (n = 4). ∗P <.05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. (E) UMAP plot three-colored to show the clusters found in the gastroid lines. There are 2 distinctive cell clusters present in the Meta1 gastroid-derived cells, Cluster A and Cluster B. Cluster C contains most of Meta4 gastroid cells. (F) Gene set enrichment analysis of the Kyoto Encyclopedia of Genes and Genomes database using the up-regulated genes in Cluster B versus Cluster A.