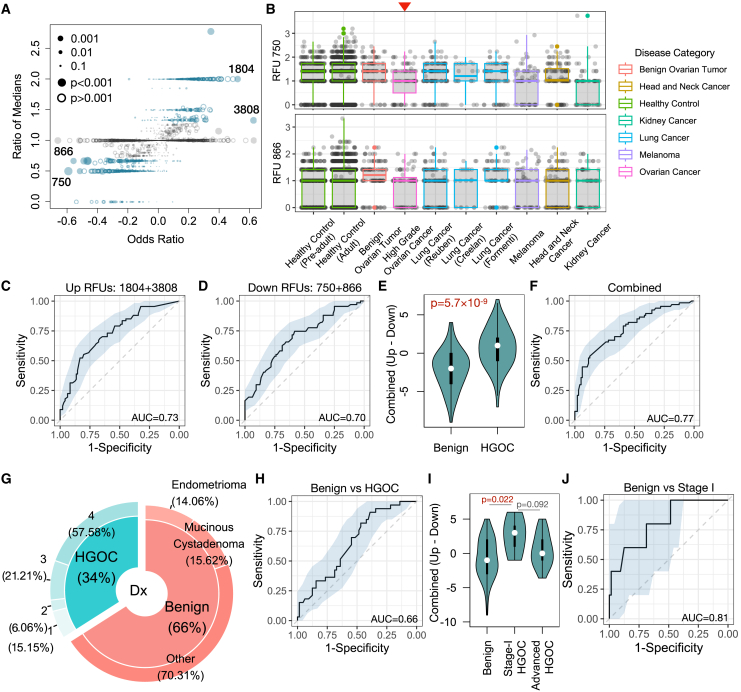
Figure 3.
Selected RFUs as biomarkers to distinguish HGOCs from benign ovarian lesions
(A) Selection criteria for the top informative RFUs. Odds ratios and ratios of medians (HGOC vs. benign) are displayed in a scatterplot. Odds ratios were calculated from the logistic regression described in Figure 2F. Blue color indicates ratio median <0.7 or >1.3.
(B) Boxplot showing the distributions of selected RFUs across multiple cancer types. Red arrow indicates ovarian cancer group. All analysis was performed using blood TCR repertoire samples from the public domain.
(C and D) Receiver operating characteristic (ROC) curves showing the prediction accuracy of up- or down-regulated RFUs to predict HGOCs against benign patients.
(E) Combination of up- and down-regulated RFUs as a joint biomarker: the OV RFU score. Statistical significance was evaluated using two-sided Wilcoxon test.
(F) Prediction accuracy of the OV RFU score illustrated by ROC curves.
(G) Donut plot showing the sample composition in the validation cohort, with total n = 97. The inner ring visualizes the percentage of patients with HGOC vs. benign patients, while the outer ring indicates the tumor stage for patients with HGOC and histological subgroups for benign patients.
(H) Performance of OV RFU score in the validation cohort.
(I) Violin plot showing the distributions of OV RFU scores across benign, stage I HGOC, and advanced HGOCs. Statistical significance was evaluated using Wilcoxon test.
(J) ROC curve for OV RFU score as a predictive biomarker for patients with stage I HGOC vs. benign patients.