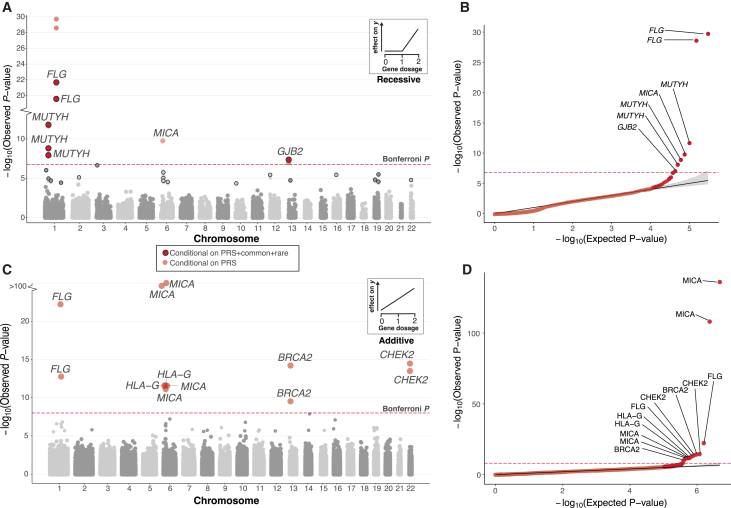
Figure 2.
Conditional recessive and additive modeling of gene copy disruption in 311 phenotypes across 176,587 participants
(A) Recessive Manhattan plot depicting log10-transformed gene-trait association p values against chromosomal location. Associations are colored red if they are Bonferroni (p < 1.68 × 10−7) significant. Transparent coloring represents the resulting p value when conditioning only on PRS, whereas solid coloring with black outline represents the p value derived after conditioning on off-chromosome PRS, nearby (500 kb) common variant association signal, and rare variants within the gene when applicable (STAR Methods). The Bonferroni significance threshold is also displayed as a red dashed line. A gene may appear multiple times if it is associated with >1 phenotype. A qualifying example of the recessive inheritance pattern is shown at the top right of the panel: disruption of both gene copies results in an effect on the phenotype.
(B) Quantile-quantile (Q-Q) plot for genes with bi-allelic damaging variants after conditioning on off-chromosome PRS. The shaded area depicts the 95% CI under the null. Gene-trait associations passing Bonferroni significance are labeled accordingly.
(C and D) Additive Manhattan plot and corresponding Q-Q plot for genes with mono- and bi-allelic damaging variants associated with at least 1 phenotype after conditioning on off-chromosome PRS when applicable (STAR Methods). No additional conditioning was performed in this analysis. Gene-trait associations are colored red if they are Bonferroni (p < 9.8 × 10−9) significant. The additive inheritance model is depicted at the top right of the panel; each affected haplotype results in a incremental effect on the phenotype.