Figure 3.
In silico permutation of genetic phase provides evidence for CH-specific effects
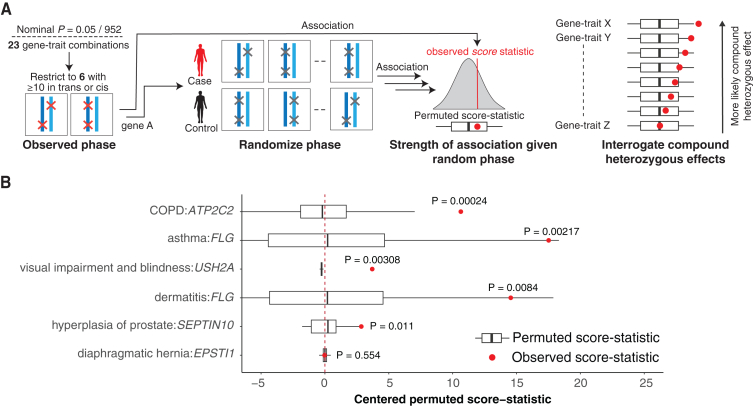
(A) Overview of the permutation pipeline. To be sufficiently powered to detect effects, we considered 5 significant (p < 0.01) gene-trait pairs from the genome-wide analysis that have at least 10 individuals harboring pLoF or damaging missense/protein-altering variants on the same haplotypes or CH carriers. Then, we shuffled CH trans and cis labels across samples and re-ran the association analysis, resulting in a null distribution of permuted score statistics corresponding to the association strength in the absence of phase information. We derive the 1-tailed empirical p value by comparing the observed score statistics with the empirical null distribution.
(B) The resulting distributions of permuted (white and black boxplots) and observed score statistic (red dot) for each gene-trait and the resulting empirical p value. p values shown in bold indicate Bonferroni significance ((p < 0.05/06 = 0.0083). Box and whisker plots display the quartiles of the empirical null distribution.