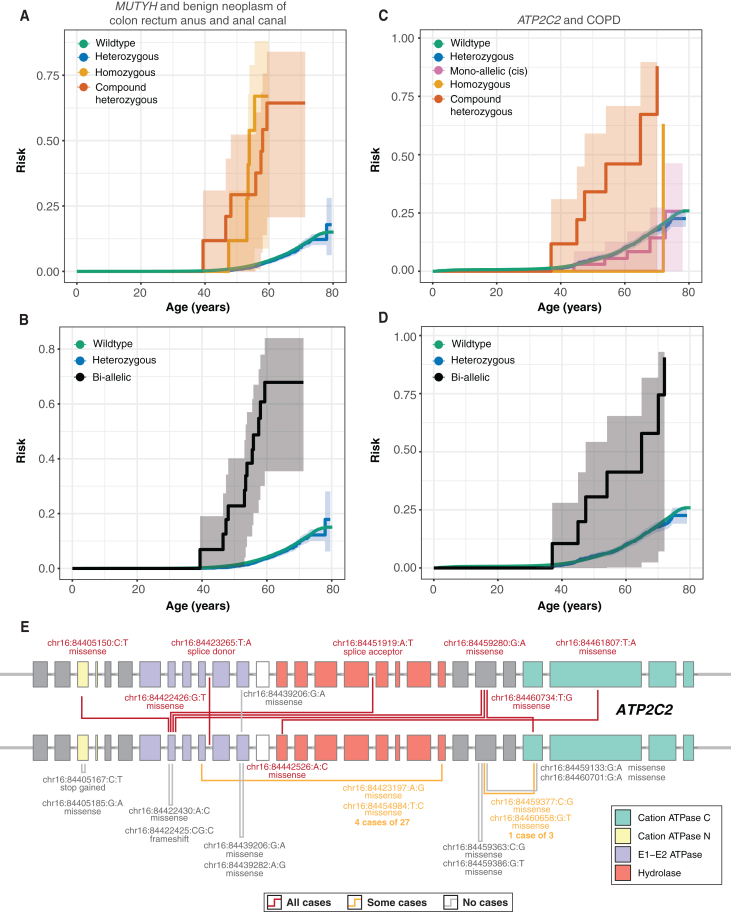
Figure 5.
Trajectories of haplotype disruption in common diseases
(A and B) Kaplan-Meier survival curves for CH (red), homozygous (orange), heterozygous carriers (blue), and single disruption of haplotypes (pink) due to pLoF or damaging missense/protein-altering mutations. Shaded regions indicate 95% confidence intervals for risk estimates. Wild types and bi-allelic variants (CH or homozygous) are shown with green and black lines, respectively. Both CH and homozygous MUTYH-variant carriers are at elevated lifetime risk of developing benign neoplasm of the colon compared to heterozygous carriers and wild types.
(C and D) Kaplan-Meier survival curves for ATP2C2 mono- and bi-allelic variant carriers. Carriers of CH variants develop COPD earlier compared to heterozygotes carriers and wild types. Moreover, individuals who harbor a single putatively disrupted haplotype due to ≥2 damaging variants develop COPD at the same frequency as heterozygotes and wild types.
(E) Gene plots for ATP2C2, displaying protein coding variants for samples that carry ≥2 pLoF or damaging missense/protein-altering variants stratified by exon or intron. CH variants, multiple variants in cis, and homozygous variants are highlighted by lines joining the positions of co-occurring variants in a sample. Lines are colored by the number of cases for the shown variant configurations, with gray lines indicating no observed samples are cases, orange lines indicating some some samples are cases, and red lines indicating that all observed samples are cases. Variants are labeled by position (GRCh38) and according to inferred consequence (missense, stop gain, splice acceptor/donor). Protein domains are highlighted accordingly.41