Figure 3.
Haplotype resolution using OGM
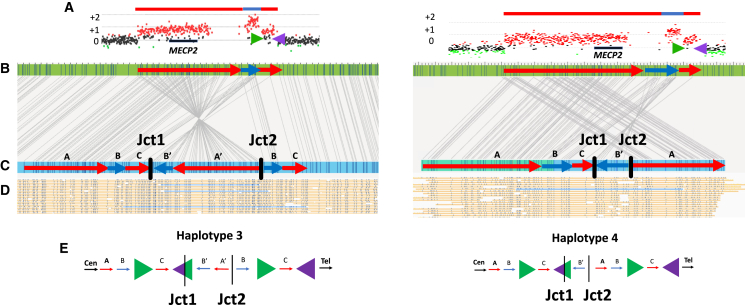
Structural haplotype determination and conformer configuration was established based upon single-molecule support through junction 1 and junction 2 in cis. Two samples in the cohort are highlighted, BAB14604 (left) and BAB15418 (right).
(A) ArrayCGH plots for each sample show a similarly sized DUP-TRP-DUP event both mediated by inverted LCRs (shown as green and purple arrows) downstream of MECP2 (black rectangle).
(B) OGM reference (green rectangle) shows in silico motifs throughout the MECP2 locus. The red arrows correspond to the duplicated segments, whereas the blue arrow corresponds to the triplicated segments. The length of the CNVs is proportional to the aCGH CNV.
(C) OGM de novo assembly from proband samples are shown in blue rectangles. Sequence motifs aligned to the reference shown as connecting gray lines enable restriction fragment genome mapping and pattern recognition. Red and blue arrows are overlayed to represent the position and orientation of each amplified genomic fragment within the DUP-TRP-DUP structure. The connection points forming junctions 1 and 2 are shown as black vertical dashed lines/bars.
(D) Single DNA molecules that span both junctions 1 and 2 are highlighted in blue, confirming that both junctions are present in cis.
(E) Hypothesized resolved haplotypes based on CNV and in cis junction analysis. Although both samples show nearly identical aCGH patterns, BAB14604 has conformer haplotype 3 and BAB15418 shows conformer haplotype 4.