Figure 4.
Refined position of TS within LCR K1/K2
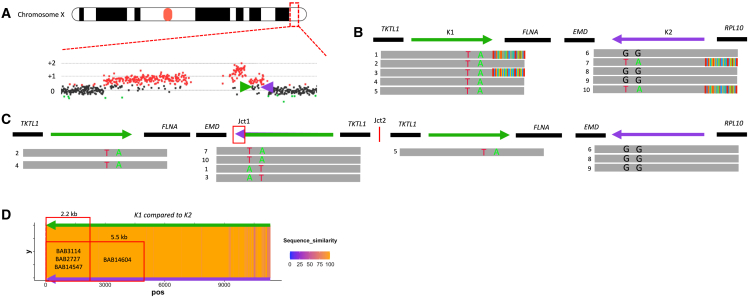
Identification of PSVs through inverted LCRs allows for a determination of the relative position of the breakpoint junction within the inverted LCRs for BAB2727.
(A) aCGH showing a DUP-TRP-DUP structure, with MECP2 locus highlighted and magnified. The inverted LCRs K1 and K2 (shown as green and purple arrows) are located flanking the terminal/3′ end duplication in the structure.
(B) Positions of K1 and K2 are shown with representative HiFi data below, highlighting sequence reads that span the region. Ancestral reads denote HiFi reads are uniquely aligned with LCR (e.g., reads 2, 4, and 5). Breakpoint reads denote HiFi reads that begin in unique sequence and show soft clipping as they exit the LCR (e.g., reads 1 and 3). PSVs are visualized in LCR K2 with the green (A nucleotide) and red (T nucleotide) positions (ChrX:153,615,342 and ChrX:153,615,645, respectively) that are found breakpoint reads in K2 and are present within points of homology in K1.
(C) Linearized structure showing the reads found within each position. The chimeric K1/K2 shows the positioning of PSVs used to refine the position of Jct1.
(D) Percentage of uniquely aligned base in slide window of 20 bp (i.e., sequence similarities were shown as a heatmap). A “hot” color, orange, denotes a 100% match, while a “cold” color, purple, denotes reduced similarity. The position of the PSV can be used to estimate the distance the replication fork proceeds before the TS to K2 occurred. Samples in this cohort could be narrowed to a 2.2- or 5.5-kb region.