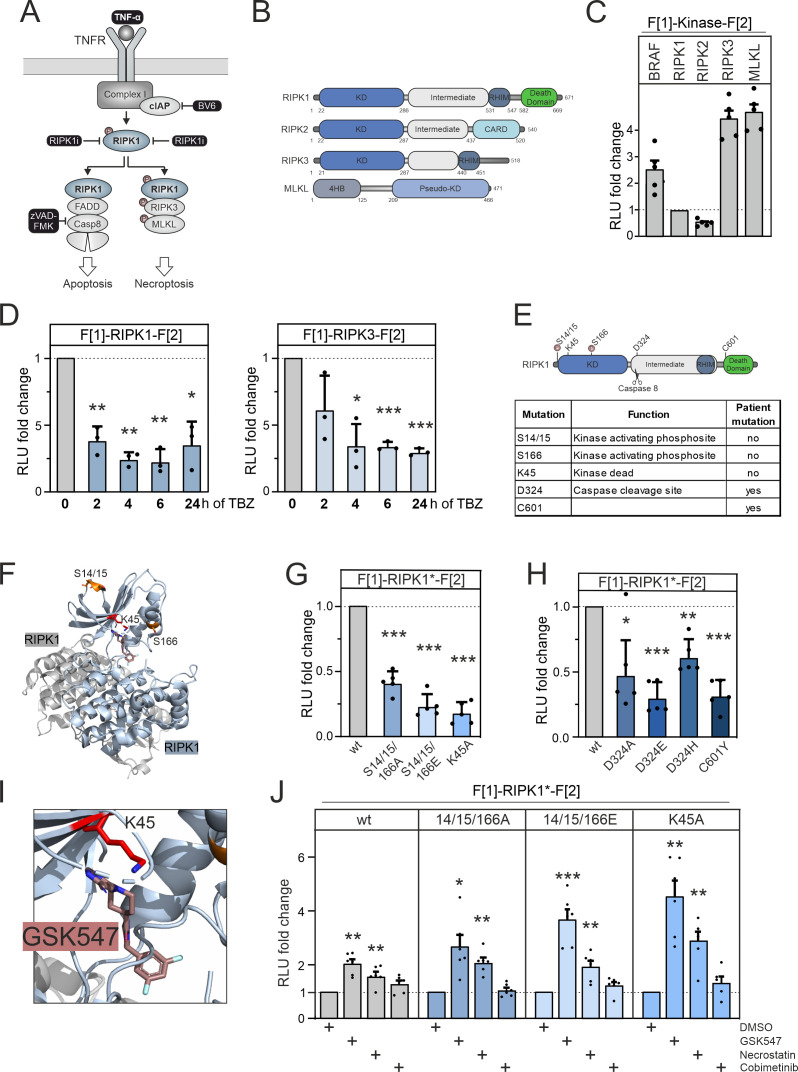
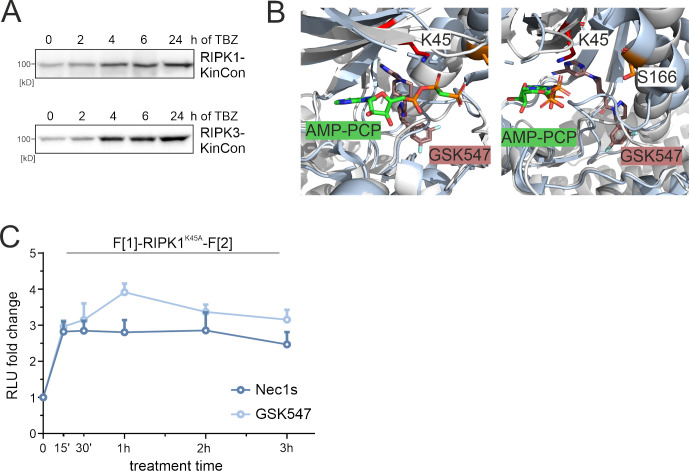
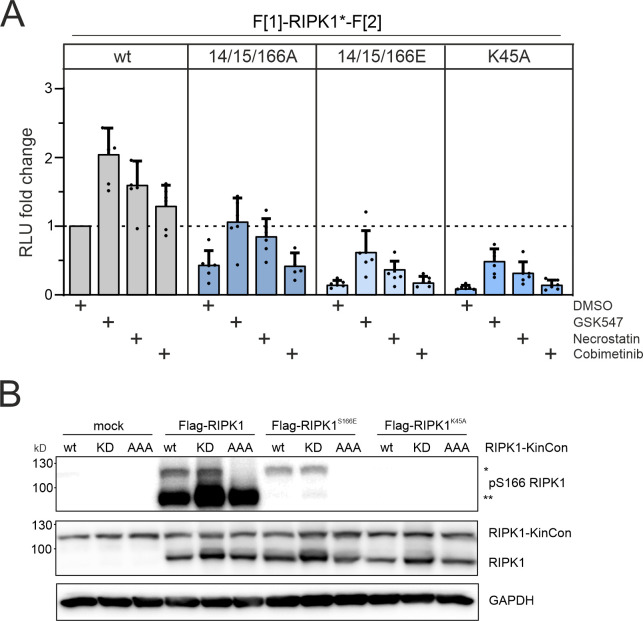
Figure 3. RIPK1 conformation dynamics.
(A) Simplified schematic representations of the activation pathways for apoptosis and necroptosis. Highlighted in black is the combination treatment termed TBZ (10 pg/ml TNFα, 10 nM BV-6, and 20 nM zVAD-FMK) that induces necroptosis. (B) Domain organization of human RIPK1 (accession number: Q13546), RIPK2 (accession number: O43353), RIPK3 (accession number: Q9Y572), and MLKL (accession number: Q8NB16). (C) Basal signals of indicated kinase conformation (KinCon) reporters following transient over-expression in HEK293T cells. Bars represent the RLU fold change relative to RIPK1 (mean ± SD, n=5 ind. experiments). (D) Time-dependent treatments using TBZ of HEK293T cells transiently expressing wild-type (wt) RIPK1 (left) and wt RIPK3 (right) KinCon reporters (expression corrected) (mean ± SD, n=3 ind. experiments). (E) Domain organization of RIPK1 displaying missense mutation sites. (F) 3D structure of RIPK1 dimers with functional mutations highlighted (PDB code: 6HHO, Wang et al., 2018). GSK547 is depicted as brown sticks. (G) KinCon reporter signals with/without mutations (S14/15/166A, S14/15/166E, K45A) were measured in a HEK293T RIPK1 knock-out (KO) cell line (expression corrected) (mean ± SEM, n=5 ind. experiments). (H) KinCon reporter signals of RIPK1 (patient loci: D324A, D324E, D324H, C601Y) were measured in HEK293T RIPK1 KO cells (expression corrected) (mean ± SD, n=5 ind. experiments). (I) 3D structure of RIPK1 with the inhibitor GSK547, which binds to an allosteric site in close proximity to the ATP-binding site (PDB code: 6HHO, Wang et al., 2018). (J) RIPK1 reporter signals with indicated mutations (described in G) upon exposure to GSK547 and Necrostatin 1 μM, and the MEKi Cobimetinib (1 μM, control experiment) or DMSO for 1 hr (mean ± SD, n=6 ind. experiments, HEK293T RIPK1 KO). Statistical significance for C–J: one-sample t-test (*p<0.05, **p<0.01, ***p<0.001).