FIGURE 1.
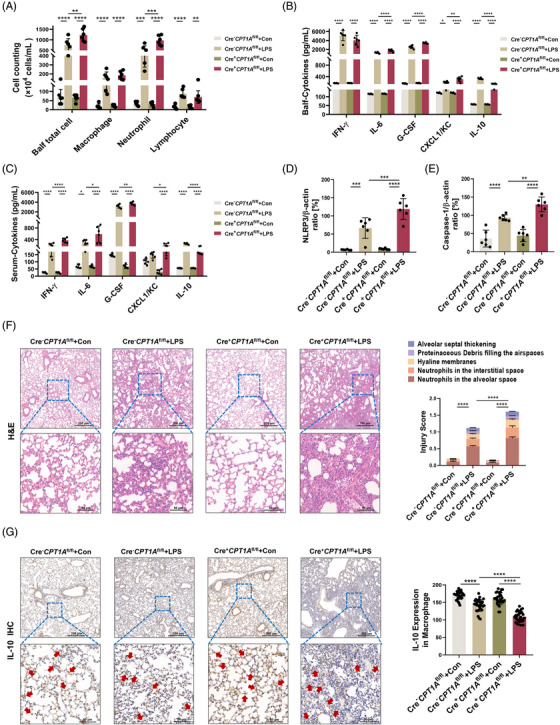
CPT1A deficiency in macrophage contributes to more severe lung injury in the ALI mouse model. (a) Cre− CPT1A fl/fl and Cre+ CPT1A fl/fl mice were administrated nasally with PBS or LPS (10 mg/kg). After 24 h, mice were euthanised and Balf was collected and estimated for total cell number, macrophage, neutrophil and lymphocyte numbers. n = 6 biologically independent samples. Balf total cell, **p = .0044, ****p < .0001. Macrophage, ****p < .0001. Neutrophil, ***p = .0002, ****p < .0001. Lymphocyte, **p = .002, ****p < .0001. (b, c) Secretion of cytokines IFN‐γ, IL‐6, G‐CSF, CXCL1/KC and IL‐10 in Balf and serum was assessed by ELISA kits. n = 6 biologically independent samples. (b) *p = .0212, **p = .0017, ****p < .0001. (c) *p = .0368 (Cre− CPT1A fl/fl+Con group vs. Cre− CPT1A fl/fl+LPS group), *p = .0116 (Cre− CPT1A fl/fl+LPS group vs. Cre+ CPT1A fl/fl+LPS group), **p = .0046, *p = .0448, ****p < .0001. (d, e) Quantitative analysis of immunoblotting assay identifying the activation of NLRP3/Caspase‐1 pathway for lung inflammation evaluation. n = 6 biologically independent samples. (d) ***p = .0002 (Cre− CPT1A fl/fl+Con group vs. Cre− CPT1A fl/fl+LPS group), ***p = .0009 (Cre− CPT1A fl/fl+LPS group vs. Cre+ CPT1A fl/fl+LPS group), ****p < .0001. (e) **p = .0086, ****p < .0001. (f) Representative pictures of H&E‐stained lung tissue sections from each group. Scale bars in the upper panel, 200 µm; scale bars in the lower panel, 50 µm. Lung injury score was determined by 5 pathophysiological features. n = 6 biologically independent samples. ****p < .0001. (g) Immunohistochemical staining and quantitative analysis of IL‐10 in Cre− CPT1A fl/fl and Cre+ CPT1A fl/fl mice. Red arrowheads indicate IL‐10‐expressed macrophage within lungs. n = 30 each group from 6 biologically independent samples. ****p < .0001. Scale bars in the upper panel, 200 µm; scale bars in the lower panel, 50 µm. Data are presented as mean ± SEM and analysed with a 95% confidence interval. p Values were calculated using one‐way ANOVA followed by Bonferroni's post hoc test.
