FIGURE 5.
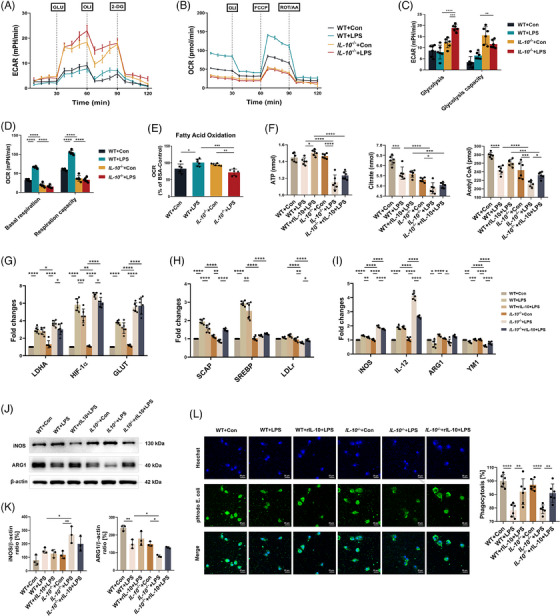
IL‐10 alters metabolic profiles and polarisation of macrophages upon LPS challenge. (a, b) ECAR (a) and OCR (b) of BMDMs from WT and IL‐10 −/− mice were measured with a Seahorse XFe96 analyser. n = 6 biologically independent samples at each data point. (c–e) The glycolysis level and glycolysis capacity (c), as well as basal respiration and Respiration capacity (d), and FAO level (e) of BMDMs from WT and IL‐10 −/− mice with or without LPS stimulation (100 ng/mL). n = 6 biologically independent samples. (c) ***p = .0004, **p = .0083, ****p < .0001. (d) ****p < .0001. (e) ***p = .0003, **p = .0029, *p = .0224. (f) After 12 h treatment with LPS (100 ng/mL) with or without rIL‐10 addition in BMDMs, generation of ATP, as well as intracellular FAO metabolites, citrate and acetyl CoA were measured. n = 6 biologically independent samples. *p = .0093, *p = .0145, ***p = .0006 (WT+rIL‐10+LPS group vs. IL‐10 −/−+rIL‐10+LPS group), ***p = .0001 (WT+Con group vs. WT+LPS group), *p = .017, ***p = .0004 (WT+rIL‐10+LPS group vs. IL‐10 −/−+rIL‐10+LPS group), ***p = .0001 (IL‐10 −/−+Con group vs. IL‐10 −/−+LPS group), ****p < .0001. (g, h) Induction of LDHA, HIF‐1α and GLUT (g), as well as SCAP, SREBP and LDLr (h) mRNA expression in WT and IL‐10 −/− BMDMs was estimated by qRT‐PCR. Data are expressed as fold change. n = 6 biologically independent samples. (g) *p = .0407 (IL‐10 −/−+LPS group vs. IL‐10 −/−+rIL‐10+LPS group), *p = .0164 (WT+LPS group vs. IL‐10 −/−+LPS group), *p = .035, **p = .0038, ***p = .0008, ****p < .0001. (h) **p = .0034, **p = .0063, *p = .0202, **p = .0061,****p < .0001. (i) Induction of iNOS, IL‐12, ARG1 and YM1 mRNA expression in WT and IL‐10 −/− BMDMs was analysed via qRT‐PCR. Data are expressed as fold change. n = 6 biologically independent samples. **p = .0018, *p = .0245, *p = .0286, **p = .0025, ***p = .0004, ****p < .0001. (j, k) Immunoblotting (j) and its quantitative analysis (k) verifying the effect of IL‐10 on iNOS and ARG1 expression for macrophage polarisation in BMDMs with or without LPS (100 ng/mL) challenge. n = 3 biologically independent samples. iNOS, *p = .0287, **p = .0052. ARG1, **p = .0011, *p = .011 (IL‐10 −/−+Con group vs. IL‐10 −/−+LPS group), *p = .0112 (WT+LPS group vs. IL‐10 −/−+LPS group). (l) The phagocytic capacity of BMDMs from WT and IL‐10 −/− mice was evaluated using the pHrod Green E. coli BioParticles through both confocal microscopy (showed in the left panel; scale bars, 20 µm) and fluorescence plate reader (showed in the right panel). n = 6 biologically independent samples. **p = .002 (WT+LPS group vs. WT +rIL‐10+LPS group), **p = .0088 (IL‐10 −/−+LPS group vs. IL‐10 −/−+rIL‐10+LPS group), ****p < .0001. Data are presented as mean ± SEM and analysed with a 95% confidence interval. p Values were calculated using one‐way ANOVA followed by Bonferroni's post hoc test.
