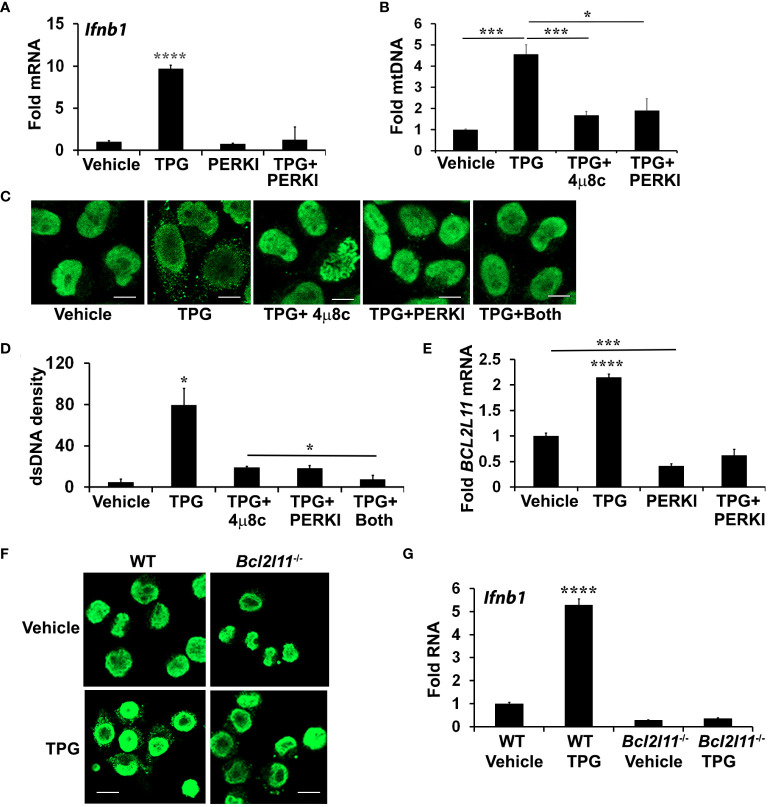
Figure 4.
PERK and Bim regulate TPG-dependent IFNB1 induction and cytoplasmic dsDNA release. (A) iMac cells were pre-treated with 10μM of PERK I (GSK2606414) for 30 min before stimulation with DMSO vehicle or 1μM of TPG for 3h. Ifnb1 mRNA was quantitated using qPCR. N=3 exp and ****p<0.001 comparing TPG with other conditions. Fold RNA is vs. vehicle control. (B) HeLa cells were pre-treated with 4μ8c or PERKI prior to stimulation with TPG for 1h. mtDNA in cytosolic fractions were quantitated by qPCR and normalized to mtDNA in whole cell extracts. N=3. *p<0.05 and ***p<0.005 in pairwise comparisons. (C) HeLa cells were treated as in (B) and dsDNA was visualized using immunofluorescence. (D) The dsDNA fluorescence densities were quantitated with ImageJ. Results are averages from 3 fields and are representative of 3 independent experiments. *p<0.05 comparing TPG with all other conditions and TPG+4μ8c with TPG+Both (E) HeLa cells were treated as in (A) and Bim (BCL2L11) mRNA quantitated by qPCR as above. ****p<0.001 for TPG vs. all other conditions and ***p<0.005 comparing vehicle only vs PERK I treated cells. (F) WT or Bim (Bcl2l11)-/- iMacs were stimulated with 1 μM TPG for 1h prior to fixation and dsDNA staining for immunoflourescence. Scale bars are 10 μm. (G) or 3h prior to harvesting for RNA extraction and qPCR. ****P-value compares TPG treated WT iMacs and either untreated cells or TPG-stimulated Bcl2l11 -/- cells. N=3.