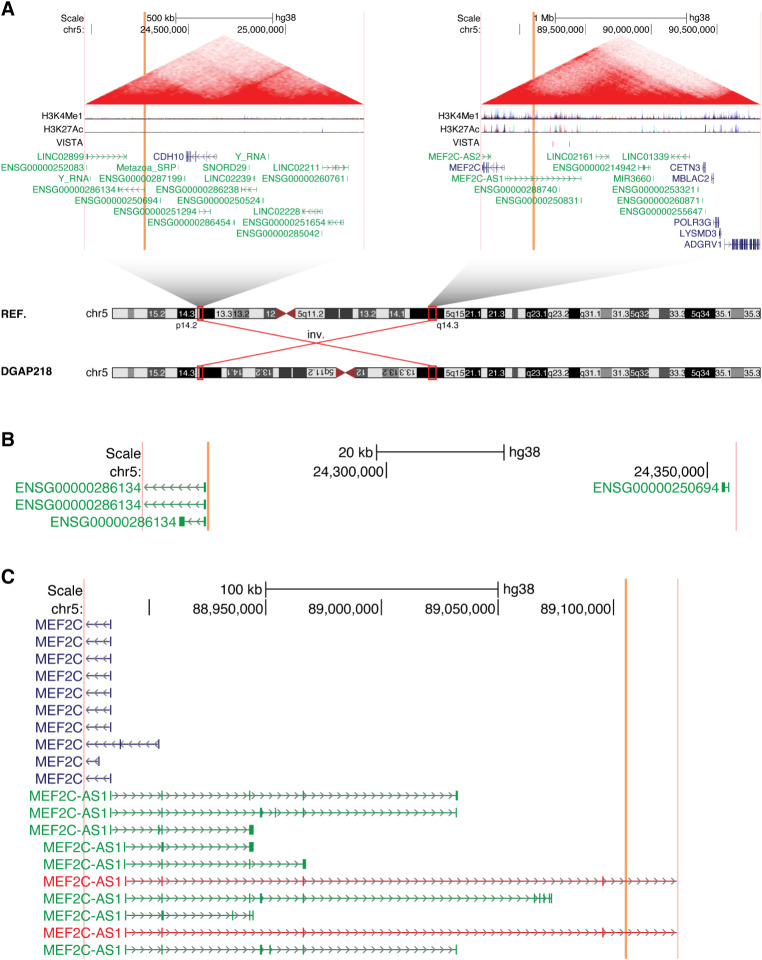
Fig. 3.
The lncRNA MEF2C-AS1 is disrupted in multiple individuals with neurological phenotypes, as shown here for DGAP218. (A) Chromosome diagrams depict the inversion between 5p14.2 and 5q14.3 in DGAP218. Above, TADs containing the breakpoints are shown, with the breakpoint positions indicated by vertical orange bars and the edges of the region shown in vertical pink bars. TAD borders were defined in (Dixon et al. 2012). Triangular contact maps display micro-C data indicating chromatin conformation (Krietenstein et al. 2020). H3K4Me1 and H3K27Ac tracks depict enhancer-associated chromatin modifications (ENCODE Project Consortium 2012). The VISTA track shows experimentally validated enhancer elements (Visel et al. 2007). Protein-coding genes are shown in blue and non-coding genes in green, with a single isoform depicted per gene. (B) Expanded view of the genomic region surrounding the 5p14.2 breakpoints in DGAP218. (C) Expanded view of the genomic region surrounding the 5q14.3 breakpoints in DGAP218. The directly disrupted lncRNA MEF2C-AS1 is highlighted in red